3GZF
 
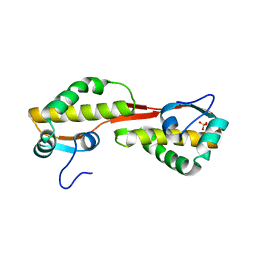 | | Structure of the C-terminal domain of nsp4 from Feline Coronavirus | | Descriptor: | Replicase polyprotein 1ab, SULFATE ION | | Authors: | Manolaridis, I, Wojdyla, J.A, Panjikar, S, Snijder, E.J, Gorbalenya, A.E, Coutard, B, Tucker, P.A. | | Deposit date: | 2009-04-07 | | Release date: | 2009-08-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.756 Å) | | Cite: | Structure of the C-terminal domain of nsp4 from feline coronavirus
Acta Crystallogr.,Sect.D, 65, 2009
|
|
1QZ8
 
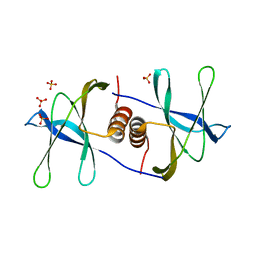 | | Crystal structure of SARS coronavirus NSP9 | | Descriptor: | SULFATE ION, polyprotein 1ab | | Authors: | Egloff, M.P, Ferron, F, Campanacci, V, Longhi, S, Rancurel, C, Dutartre, H, Snijder, E.J, Gorbalenya, A.E, Cambillau, C, Canard, B. | | Deposit date: | 2003-09-16 | | Release date: | 2004-02-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The severe acute respiratory syndrome-coronavirus replicative protein nsp9 is a single-stranded RNA-binding subunit unique in the RNA virus world.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1MBM
 
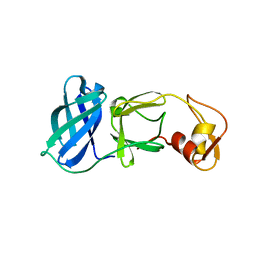 | | NSP4 proteinase from Equine Arteritis Virus | | Descriptor: | chymotrypsin-like serine protease | | Authors: | Barrette-Ng, I.H, Ng, K.K.-S, Mark, B.L, van Aken, D, Cherney, M.M, Garen, C, Kolodenko, Y, Gorbalenya, A.E, Snijder, E.J, James, M.N.G. | | Deposit date: | 2002-08-03 | | Release date: | 2002-10-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Arterivirus nsp4: the smallest chymotrypsin-like proteinase with an alpha/beta C-terminal extension and alternate conformations of the oxyanion hole
J.Biol.Chem., 277, 2002
|
|
3MP2
 
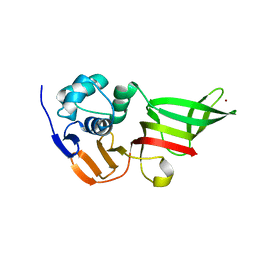 | |
2L8K
 
 | | NMR Structure of the Arterivirus nonstructural protein 7 alpha (nsp7 alpha) | | Descriptor: | Non-structural protein 7 | | Authors: | Conte, M.R, Gaudin, C, Manolaridis, I, Tucker, P.W, Kelly, G. | | Deposit date: | 2011-01-19 | | Release date: | 2011-07-06 | | Last modified: | 2023-02-08 | | Method: | SOLUTION NMR | | Cite: | Structure and Genetic Analysis of the Arterivirus Nonstructural Protein 7{alpha}.
J.Virol., 85, 2011
|
|
6FV1
 
 | | Structure of human coronavirus NL63 main protease in complex with the alpha-ketoamide (S)-N-((S)-4-(benzylamino)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl)butan-2-yl)-2-cinnamamido-4-methylpentanamide (cinnamoyl-leucine-GlnLactam-CO-CO-NH-benzyl) | | Descriptor: | (2~{S})-4-methyl-~{N}-[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]-2-[[(~{E})-3-phenylprop-2-enoyl]amino]pentanamide, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Zhang, L, Hilgenfeld, R. | | Deposit date: | 2018-02-28 | | Release date: | 2019-03-20 | | Last modified: | 2021-04-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Alpha-ketoamides as broad-spectrum inhibitors of coronavirus and enterovirus replication Structure-based design, synthesis, and activity assessment.
J.Med.Chem., 2020
|
|
6FV2
 
 | |
3JZT
 
 | | Structure of a cubic crystal form of X (ADRP) domain from FCoV with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, CHLORIDE ION, SODIUM ION, ... | | Authors: | Wojdyla, J.A, Manolaridis, I, Tucker, P.A. | | Deposit date: | 2009-09-24 | | Release date: | 2010-01-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.91 Å) | | Cite: | Structure of the X (ADRP) domain of nsp3 from feline coronavirus
Acta Crystallogr.,Sect.D, 65, 2009
|
|
4N0N
 
 | | Crystal structure of Arterivirus nonstructural protein 10 (helicase) | | Descriptor: | MAGNESIUM ION, Replicase polyprotein 1ab, SULFATE ION, ... | | Authors: | Deng, Z, Chen, Z. | | Deposit date: | 2013-10-02 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the regulatory function of a complex zinc-binding domain in a replicative arterivirus helicase resembling a nonsense-mediated mRNA decay helicase.
Nucleic Acids Res., 42, 2014
|
|
4N0O
 
 | | Complex structure of Arterivirus nonstructural protein 10 (helicase) with DNA | | Descriptor: | CALCIUM ION, DNA, Replicase polyprotein 1ab, ... | | Authors: | Deng, Z, Chen, Z. | | Deposit date: | 2013-10-02 | | Release date: | 2014-01-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis for the regulatory function of a complex zinc-binding domain in a replicative arterivirus helicase resembling a nonsense-mediated mRNA decay helicase.
Nucleic Acids Res., 42, 2014
|
|
4RF1
 
 | | Crystal structure of the Middle-East respiratory syndrome coronavirus papain-like protease in complex with ubiquitin (space group P63) | | Descriptor: | 3-AMINOPROPANE, ORF1ab protein, S-1,2-PROPANEDIOL, ... | | Authors: | Bailey-Elkin, B.A, Johnson, G.G, Mark, B.L. | | Deposit date: | 2014-09-24 | | Release date: | 2014-10-22 | | Last modified: | 2015-01-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of the Middle East Respiratory Syndrome Coronavirus (MERS-CoV) Papain-like Protease Bound to Ubiquitin Facilitates Targeted Disruption of Deubiquitinating Activity to Demonstrate Its Role in Innate Immune Suppression.
J.Biol.Chem., 289, 2014
|
|
4REZ
 
 | | Crystal structure of the Middle-East respiratory syndrome coronavirus papain-like protease | | Descriptor: | ORF1ab protein, S-1,2-PROPANEDIOL, ZINC ION | | Authors: | Bailey-Elkin, B.A, Johnson, G.G, Mark, B.L. | | Deposit date: | 2014-09-24 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Middle East Respiratory Syndrome Coronavirus (MERS-CoV) Papain-like Protease Bound to Ubiquitin Facilitates Targeted Disruption of Deubiquitinating Activity to Demonstrate Its Role in Innate Immune Suppression.
J.Biol.Chem., 289, 2014
|
|
4RF0
 
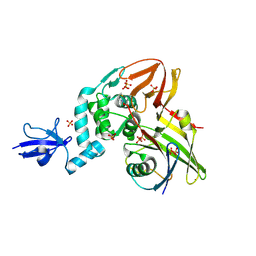 | | Crystal structure of the Middle-East respiratory syndrome coronavirus papain-like protease in complex with ubiquitin (space group P6522) | | Descriptor: | 3-AMINOPROPANE, ORF1ab protein, SULFATE ION, ... | | Authors: | Bailey-Elkin, B.A, Johnson, G.G, Mark, B.L. | | Deposit date: | 2014-09-24 | | Release date: | 2014-10-22 | | Last modified: | 2015-01-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Middle East Respiratory Syndrome Coronavirus (MERS-CoV) Papain-like Protease Bound to Ubiquitin Facilitates Targeted Disruption of Deubiquitinating Activity to Demonstrate Its Role in Innate Immune Suppression.
J.Biol.Chem., 289, 2014
|
|
3EJF
 
 | |
3EJG
 
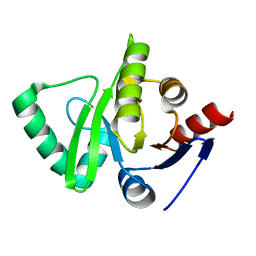 | | Crystal structure of HCoV-229E X-domain | | Descriptor: | Non-structural protein 3 | | Authors: | Piotrowski, Y, Hansen, G, Hilgenfeld, R. | | Deposit date: | 2008-09-18 | | Release date: | 2008-09-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structures of the X-domains of a Group-1 and a Group-3 coronavirus reveal that ADP-ribose-binding may not be a conserved property.
Protein Sci., 18, 2009
|
|
3EKE
 
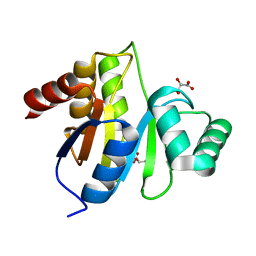 | | Crystal structure of IBV X-domain at pH 5.6 | | Descriptor: | L(+)-TARTARIC ACID, Non-structural protein 3 | | Authors: | Piotrowski, Y, Hansen, G, Hilgenfeld, R. | | Deposit date: | 2008-09-19 | | Release date: | 2008-09-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the X-domains of a Group-1 and a Group-3 coronavirus reveal that ADP-ribose-binding may not be a conserved property.
Protein Sci., 18, 2009
|
|
3ETI
 
 | |
3EW5
 
 | | Structure of the tetragonal crystal form of X (ADRP) domain from FCoV | | Descriptor: | CHLORIDE ION, SN-GLYCEROL-1-PHOSPHATE, SULFATE ION, ... | | Authors: | Wojdyla, J.A, Manolaridis, I, Tucker, P.A. | | Deposit date: | 2008-10-14 | | Release date: | 2009-10-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the X (ADRP) domain of nsp3 from feline coronavirus
Acta Crystallogr.,Sect.D, 65, 2009
|
|
4IUM
 
 | | Equine arteritis virus papain-like protease 2 (PLP2) covalently bound to ubiquitin | | Descriptor: | GLYCEROL, Ubiquitin, ZINC ION, ... | | Authors: | Bailey-Elkin, B.A, James, T.W, Mark, B.L. | | Deposit date: | 2013-01-21 | | Release date: | 2013-02-13 | | Last modified: | 2013-03-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Deubiquitinase function of arterivirus papain-like protease 2 suppresses the innate immune response in infected host cells.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5N19
 
 | |
5N5O
 
 | |
5NH0
 
 | |
5NFS
 
 | |
1LVO
 
 | | Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,4-DIETHYLENE DIOXIDE, Replicase, ... | | Authors: | Anand, K, Palm, G.J, Mesters, J.R, Siddell, S.G, Ziebuhr, J, Hilgenfeld, R. | | Deposit date: | 2002-05-29 | | Release date: | 2002-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain.
EMBO J., 21, 2002
|
|
