1ESK
 
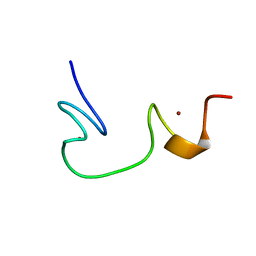 | | SOLUTION STRUCTURE OF NCP7 FROM HIV-1 | | Descriptor: | GAG POLYPROTEIN, ZINC ION | | Authors: | Morellet, N, Demene, H, Teilleux, V, Huynh-Dinh, T, de Rocquigny, H, Fournie-Zaluski, M.-C, Roques, B.P. | | Deposit date: | 2000-04-10 | | Release date: | 2000-04-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of (12-53)NCp7 of HIV-1
To be Published
|
|
1QF1
 
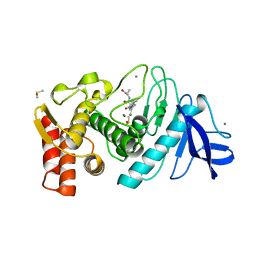 | | THERMOLYSIN (E.C.3.4.24.27) COMPLEXED WITH (2-SULPHANYLHEPTANOYL)-PHE-ALA. PARAMETERS FOR ZN-BIDENTATION OF MERCAPTOACYLDIPEPTIDES IN METALLOENDOPEPTIDASE | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, PROTEIN (THERMOLYSIN), ... | | Authors: | Gaucher, J.-F, Selkti, M, Tiraboschi, G, Prange, T, Roques, B.P, Tomas, A, Fournie-Zaluski, M.C. | | Deposit date: | 1999-04-06 | | Release date: | 1999-12-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of alpha-mercaptoacyldipeptides in the thermolysin active site: structural parameters for a Zn monodentation or bidentation in metalloendopeptidases.
Biochemistry, 38, 1999
|
|
1QF2
 
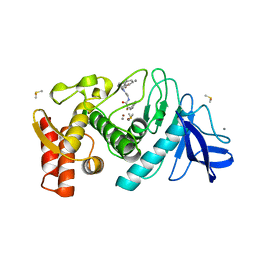 | | THERMOLYSIN (E.C.3.4.24.27) COMPLEXED WITH (2-SULPHANYL-3-PHENYLPROPANOYL)-GLY-(5-PHENYLPROLINE). PARAMETERS FOR ZN-MONODENTATION OF MERCAPTOACYLDIPEPTIDES IN METALLOENDOPEPTIDASE | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, PROTEIN (THERMOLYSIN), ... | | Authors: | Gaucher, J.-F, Selkti, M, Tiraboschi, G, Prange, T, Roques, B.P, Tomas, A, Fournie-Zaluski, M.C. | | Deposit date: | 1999-04-06 | | Release date: | 1999-12-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structures of alpha-mercaptoacyldipeptides in the thermolysin active site: structural parameters for a Zn monodentation or bidentation in metalloendopeptidases.
Biochemistry, 38, 1999
|
|
1QF0
 
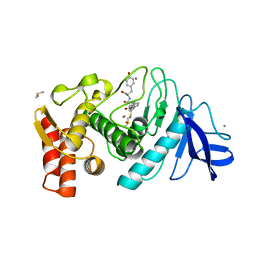 | | THERMOLYSIN (E.C.3.4.24.27) COMPLEXED WITH (2-SULPHANYL-3-PHENYLPROPANOYL)-PHE-TYR. PARAMETERS FOR ZN-BIDENTATION OF MERCAPTOACYLDIPEPTIDES IN METALLOENDOPEPTIDASE | | Descriptor: | (2-SULFANYL-3-PHENYLPROPANOYL)-PHE-TYR, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Gaucher, J.-F, Selkti, M, Tiraboschi, G, Prange, T, Roques, B.P, Tomas, A, Fournie-Zaluski, M.C. | | Deposit date: | 1999-04-06 | | Release date: | 1999-12-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of alpha-mercaptoacyldipeptides in the thermolysin active site: structural parameters for a Zn monodentation or bidentation in metalloendopeptidases.
Biochemistry, 38, 1999
|
|
2R59
 
 | | Leukotriene A4 hydrolase complexed with inhibitor RB3041 | | Descriptor: | ACETIC ACID, Leukotriene A-4 hydrolase, N-{(2S)-3-[(R)-[(1R)-1-amino-2-phenylethyl](hydroxy)phosphoryl]-2-benzylpropanoyl}-L-phenylalanine, ... | | Authors: | Tholander, F, Haeggstrom, J.Z, Thunnissen, M, Muroya, A, Roques, B.P, Fournie-Zaluski, M.C. | | Deposit date: | 2007-09-03 | | Release date: | 2008-08-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure-based dissection of the active site chemistry of leukotriene a4 hydrolase: implications for m1 aminopeptidases and inhibitor design.
Chem.Biol., 15, 2008
|
|
2JZW
 
 | | How the HIV-1 nucleocapsid protein binds and destabilises the (-)primer binding site during reverse transcription | | Descriptor: | DNA (5'-D(*DGP*DTP*DCP*DCP*DCP*DTP*DGP*DTP*DTP*DCP*DGP*DGP*DGP*DC)-3'), HIV-1 nucleocapsid protein NCp7(12-55), ZINC ION | | Authors: | Bourbigot, S, Ramalanjaona, N, Salgado, G.F.J, Mely, Y, Roques, B.P, Bouaziz, S, Morellet, N. | | Deposit date: | 2008-01-21 | | Release date: | 2009-01-13 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | How the HIV-1 nucleocapsid protein binds and destabilises the (-)primer binding site during reverse transcription.
J.Mol.Biol., 383, 2008
|
|
1A6B
 
 | | NMR STRUCTURE OF THE COMPLEX BETWEEN THE ZINC FINGER PROTEIN NCP10 OF MOLONEY MURINE LEUKEMIA VIRUS AND A SEQUENCE OF THE PSI-PACKAGING DOMAIN OF HIV-1, 20 STRUCTURES | | Descriptor: | DNA (5'-D(*AP*CP*GP*CP*C)-3'), ZINC FINGER PROTEIN NCP10, ZINC ION | | Authors: | Schueler, W, Dong, C.-Z, Wecker, K, Roques, B.P. | | Deposit date: | 1998-02-23 | | Release date: | 1999-08-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the complex between the zinc finger protein NCp10 of Moloney murine leukemia virus and the single-stranded pentanucleotide d(ACGCC): comparison with HIV-NCp7 complexes.
Biochemistry, 38, 1999
|
|
1U57
 
 | | NMR structure of the (345-392)Gag sequence from HIV-1 | | Descriptor: | Gag polyprotein | | Authors: | Morellet, N, Druillennec, S, Lenoir, C, Bouaziz, S, Roques, B.P. | | Deposit date: | 2004-07-27 | | Release date: | 2005-02-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Helical structure determined by NMR of the HIV-1 (345-392)Gag sequence, surrounding p2: Implications for particle assembly and RNA packaging
Protein Sci., 14, 2005
|
|
1VPC
 
 | | C-TERMINAL DOMAIN (52-96) OF THE HIV-1 REGULATORY PROTEIN VPR, NMR, 1 STRUCTURE | | Descriptor: | VPR PROTEIN | | Authors: | Schueler, W, De Rocquigny, H, Baudat, Y, Sire, J, Roques, B.P. | | Deposit date: | 1998-02-20 | | Release date: | 1999-03-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the (52-96) C-terminal domain of the HIV-1 regulatory protein Vpr: molecular insights into its biological functions.
J.Mol.Biol., 285, 1999
|
|
1DP7
 
 | | COCRYSTAL STRUCTURE OF RFX-DBD IN COMPLEX WITH ITS COGNATE X-BOX BINDING SITE | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DNA (5'-D(*CP*GP*(BRU)P*TP*AP*CP*CP*AP*(BRU)P*GP*GP*TP*AP*AP*CP*G)-3'), ... | | Authors: | Gajiwala, K.S, Chen, H, Cornille, F, Roques, B.P, Reith, W, Mach, B, Burley, S.K. | | Deposit date: | 1999-12-23 | | Release date: | 2000-03-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the winged-helix protein hRFX1 reveals a new mode of DNA binding.
Nature, 403, 2000
|
|
1M8L
 
 | |
1Z9G
 
 | | Crystal Structure Analysis of Thermolysin Complexed with the Inhibitor (R)-retro-thiorphan | | Descriptor: | (R)-RETRO-THIORPHAN, CALCIUM ION, Thermolysin, ... | | Authors: | Roderick, S.L, Fournie-Zaluski, M.C, Roques, B.P, Matthews, B.W. | | Deposit date: | 2005-04-01 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Thiorphan and retro-thiorphan display equivalent interactions when bound to crystalline thermolysin
Biochemistry, 28, 1989
|
|
1Q3Y
 
 | | NMR structure of the Cys28His mutant (D form) of the nucleocapsid protein NCp7 of HIV-1. | | Descriptor: | GAG protein, ZINC ION | | Authors: | Ramboarina, S, Druillennec, S, Morellet, N, Bouaziz, S, Roques, B.P. | | Deposit date: | 2003-08-01 | | Release date: | 2004-09-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Target specificity of human immunodeficiency virus type 1 NCp7 requires an intact conformation of its CCHC N-terminal zinc finger.
J.Virol., 78, 2004
|
|
1Q3Z
 
 | | NMR structure of the Cys28His mutant (E form) of the nucleocapsid protein NCp7 of HIV-1. | | Descriptor: | GAG protein, ZINC ION | | Authors: | Ramboarina, S, Druillennec, S, Morellet, N, Bouaziz, S, Roques, B.P. | | Deposit date: | 2003-08-01 | | Release date: | 2004-09-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Target specificity of human immunodeficiency virus type 1 NCp7 requires an intact conformation of its CCHC N-terminal zinc finger.
J.Virol., 78, 2004
|
|
1ZDP
 
 | | Crystal Structure Analysis of Thermolysin Complexed with the Inhibitor (S)-thiorphan | | Descriptor: | (2-MERCAPTOMETHYL-3-PHENYL-PROPIONYL)-GLYCINE, CALCIUM ION, Thermolysin, ... | | Authors: | Roderick, S.L, Fournie-Zaluski, M.C, Roques, B.P, Matthews, B.W. | | Deposit date: | 2005-04-14 | | Release date: | 2005-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Thiorphan and retro-thiorphan display equivalent interactions when bound to crystalline thermolysin
Biochemistry, 28, 1989
|
|
1X9V
 
 | | Dimeric structure of the C-terminal domain of Vpr | | Descriptor: | VPR protein | | Authors: | Bourbigot, S, Beltz, H, Denis, J, Morellet, N, Roques, B.P, Mely, Y, Bouaziz, S. | | Deposit date: | 2004-08-24 | | Release date: | 2005-06-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The C-terminal domain of the HIV-1 regulatory protein Vpr adopts an antiparallel dimeric structure in solution via its leucine-zipper-like domain
Biochem.J., 387, 2005
|
|
2A51
 
 | | Structure of the (13-51) domain of the nucleocapsid protein NCp8 from SIVlhoest | | Descriptor: | ZINC ION, nucleocapsid protein | | Authors: | Morellet, N, Meudal, H, Bouaziz, S, Roques, B.P. | | Deposit date: | 2005-06-30 | | Release date: | 2006-06-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the zinc finger domain encompassing residues 13-51 of the nucleocapsid protein from simian immunodeficiency virus
Biochem.J., 393, 2006
|
|
154D
 
 | | DNA DISTORTION IN BIS-INTERCALATED COMPLEXES | | Descriptor: | BIS-(N-ETHYLPYRIDINIUM-(3-METHOXYCARBAZOLE))HEXANE-1,6-DIAMINE, DNA (5'-D(*(CBR)P*GP*CP*G)-3') | | Authors: | Peek, M.E, Lipscomb, L.A, Bertrand, J.A, Gao, Q, Roques, B.P, Garbay-Jaureguiberry, C, Williams, L.D. | | Deposit date: | 1993-12-14 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | DNA distortion in bis-intercalated complexes.
Biochemistry, 33, 1994
|
|
1CEU
 
 | |
1BJ6
 
 | | 1H NMR OF (12-53) NCP7/D(ACGCC) COMPLEX, 10 STRUCTURES | | Descriptor: | DNA (5'-D(*AP*CP*GP*CP*C)-3'), NUCLEOCAPSID PROTEIN 7, ZINC ION | | Authors: | Demene, H, Morellet, N, Teilleux, V, Huynh-Dinh, T, De Rocquigny, H, Fournie-Zaluski, M.C, Roques, B.P. | | Deposit date: | 1998-07-03 | | Release date: | 1999-02-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the complex between the HIV-1 nucleocapsid protein NCp7 and the single-stranded pentanucleotide d(ACGCC).
J.Mol.Biol., 283, 1998
|
|
1R1I
 
 | | STRUCTURAL ANALYSIS OF NEPRILYSIN WITH VARIOUS SPECIFIC AND POTENT INHIBITORS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Neprilysin, ZINC ION, ... | | Authors: | Oefner, C, Roques, B.P, Fournie-Zaluski, M.C, Dale, G.E. | | Deposit date: | 2003-09-24 | | Release date: | 2004-09-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis of neprilysin with various specific and potent inhibitors.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1R1J
 
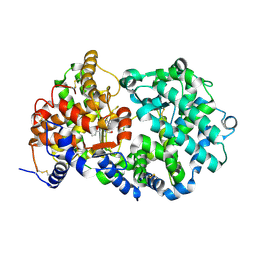 | | STRUCTURAL ANALYSIS OF NEPRILYSIN WITH VARIOUS SPECIFIC AND POTENT INHIBITORS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-(3-PHENYL-2-SULFANYLPROPANOYL)PHENYLALANYLALANINE, Neprilysin, ... | | Authors: | Oefner, C, Roques, B.P, Fournie-Zaluski, M.C, Dale, G.E. | | Deposit date: | 2003-09-24 | | Release date: | 2004-09-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural analysis of neprilysin with various specific and potent inhibitors.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1R1H
 
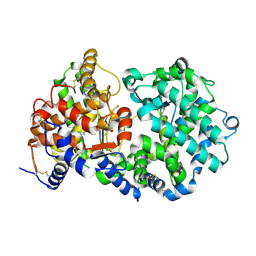 | | STRUCTURAL ANALYSIS OF NEPRILYSIN WITH VARIOUS SPECIFIC AND POTENT INHIBITORS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-[3-[(1-AMINOETHYL)(HYDROXY)PHOSPHORYL]-2-(1,1'-BIPHENYL-4-YLMETHYL)PROPANOYL]ALANINE, Neprilysin, ... | | Authors: | Oefner, C, Roques, B.P, Fournie-Zaluski, M.C, Dale, G.E. | | Deposit date: | 2003-09-24 | | Release date: | 2004-09-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis of neprilysin with various specific and potent inhibitors.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1AZE
 
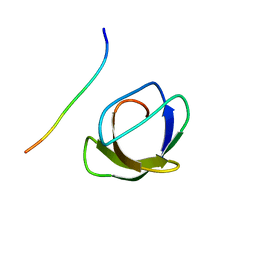 | | NMR STRUCTURE OF THE COMPLEX BETWEEN THE C32S-Y7V MUTANT OF THE NSH3 DOMAIN OF GRB2 WITH A PEPTIDE FROM SOS, 10 STRUCTURES | | Descriptor: | GRB2, SOS | | Authors: | Vidal, M, Gincel, E, Goudreau, N, Cornille, F, Parker, F, Duchesne, M, Tocque, B, Garbay, C, Roques, B.P. | | Deposit date: | 1997-11-17 | | Release date: | 1999-05-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular and cellular analysis of Grb2 SH3 domain mutants: interaction with Sos and dynamin.
J.Mol.Biol., 290, 1999
|
|
1ESX
 
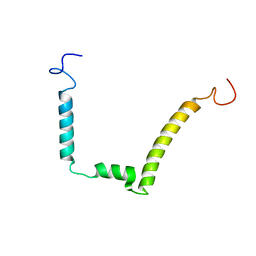 | | 1H, 15N AND 13C STRUCTURE OF THE HIV-1 REGULATORY PROTEIN VPR : COMPARISON WITH THE N-AND C-TERMINAL DOMAIN STRUCTURE, (1-51)VPR AND (52-96)VPR | | Descriptor: | VPR PROTEIN | | Authors: | Wecker, K, Morellet, N, Bouaziz, S, Roques, B. | | Deposit date: | 2000-04-11 | | Release date: | 2001-04-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the HIV-1 regulatory protein Vpr in H2O/trifluoroethanol. Comparison with the Vpr N-terminal (1-51) and C-terminal (52-96) domains.
Eur.J.Biochem., 269, 2002
|
|
