5XK4
 
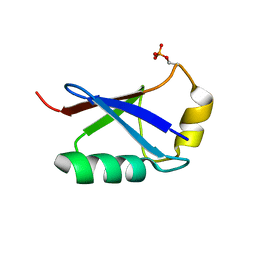 | | Retracted state of S65-phosphorylated ubiquitin | | Descriptor: | Polyubiquitin-B | | Authors: | Dong, X, Gong, Z, Qin, L.Y, Ran, M.L, Zhang, C.L, Liu, K, Liu, Z, Zhang, W.P, Tang, C. | | Deposit date: | 2017-05-05 | | Release date: | 2017-06-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Ubiquitin S65 phosphorylation engenders a pH-sensitive conformational switch
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5XK5
 
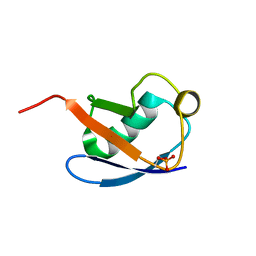 | | Relaxed state of S65-phosphorylated ubiquitin | | Descriptor: | Polyubiquitin-B | | Authors: | Xu, D, Zhou, G, Qin, L.Y, Ran, M.L, Zhang, C.L, Liu, K, Liu, Z, Zhang, W.P, Tang, C. | | Deposit date: | 2017-05-05 | | Release date: | 2017-06-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Ubiquitin S65 phosphorylation engenders a pH-sensitive conformational switch
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1NE7
 
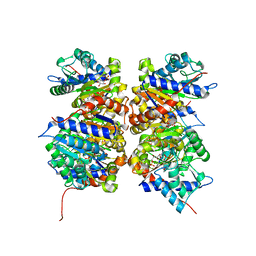 | | HUMAN GLUCOSAMINE-6-PHOSPHATE DEAMINASE ISOMERASE AT 1.75 A RESOLUTION COMPLEXED WITH N-ACETYL-GLUCOSAMINE-6-PHOSPHATE AND 2-DEOXY-2-AMINO-GLUCITOL-6-PHOSPHATE | | Descriptor: | 2-DEOXY-2-AMINO GLUCITOL-6-PHOSPHATE, 2-acetamido-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, Glucosamine-6-phosphate isomerase, ... | | Authors: | Arreola, R, Valderrama, B, Morante, M.L, Horjales, E. | | Deposit date: | 2002-12-10 | | Release date: | 2003-09-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Two mammalian glucosamine-6-phosphate deaminases: a structural and genetic study.
Febs Lett., 551, 2003
|
|
2KGB
 
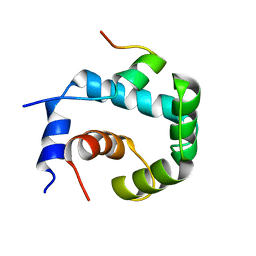 | | NMR solution of the regulatory domain cardiac F77W-Troponin C in complex with the cardiac Troponin I 144-163 switch peptide | | Descriptor: | CALCIUM ION, Troponin C, slow skeletal and cardiac muscles, ... | | Authors: | Mercier, P, Julien, O, Crane, M.L, Sykes, B.D. | | Deposit date: | 2009-03-07 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The effect of the cosolvent trifluoroethanol on a tryptophan side chain orientation in the hydrophobic core of troponin C.
Protein Sci., 18, 2009
|
|
1DEY
 
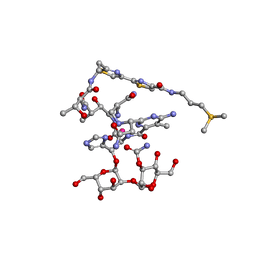 | |
2N90
 
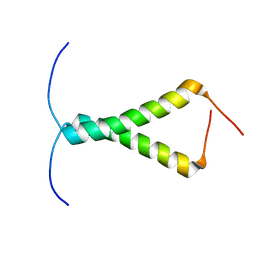 | |
5XEI
 
 | |
6I1O
 
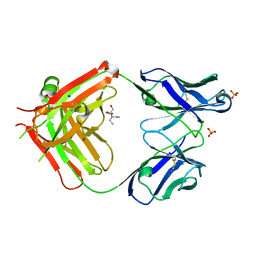 | | Fab fragment of an antibody selective for wild-type alpha-1-antitrypsin | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FAB 2H2 heavy chain, FAB 2H2 light chain, ... | | Authors: | Laffranchi, M, Elliston, E.L.K, Miranda, E, Perez, J, Fra, A, Lomas, D.A, Irving, J.A. | | Deposit date: | 2018-10-29 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Intrahepatic heteropolymerization of M and Z alpha-1-antitrypsin.
JCI Insight, 5, 2020
|
|
6I3Z
 
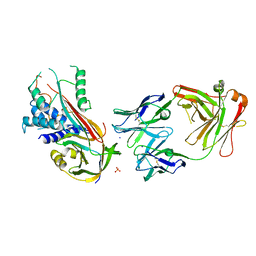 | | Fab fragment of an antibody selective for wild-type alpha-1-antitrypsin in complex with its antigen | | Descriptor: | Alpha-1-antitrypsin, Fab 2H2 heavy chain, Fab 2H2 light chain, ... | | Authors: | Laffranchi, M, Elliston, E.L.K, Miranda, E, Perez, J, Jagger, A.M, Fra, A, Lomas, D.A, Irving, J.A. | | Deposit date: | 2018-11-08 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Intrahepatic heteropolymerization of M and Z alpha-1-antitrypsin.
JCI Insight, 5, 2020
|
|
7O05
 
 | |
7O36
 
 | |
7O35
 
 | |
5NMO
 
 | | Structure of the Bacillus subtilis Smc Joint domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, Chromosome partition protein Smc,Chromosome partition protein Smc, ... | | Authors: | Diebold-Durand, M.-L, Basquin, J, Gruber, S. | | Deposit date: | 2017-04-06 | | Release date: | 2017-06-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structure of Full-Length SMC and Rearrangements Required for Chromosome Organization.
Mol. Cell, 67, 2017
|
|
5NNV
 
 | |
5XG2
 
 | |
5XG3
 
 | | Crystal structure of the ATPgS-engaged Smc head domain with an extended coiled coil bound to the C-terminal domain of ScpA derived from Bacillus subtilis | | Descriptor: | COBALT (II) ION, Chromosome partition protein Smc, MAGNESIUM ION, ... | | Authors: | Shin, H.-C, Lee, H, Oh, B.-H. | | Deposit date: | 2017-04-11 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of Full-Length SMC and Rearrangements Required for Chromosome Organization
Mol. Cell, 67, 2017
|
|
5XNS
 
 | |
5FRH
 
 | | Solution structure of oxidised RsrA | | Descriptor: | ANTI-SIGMA FACTOR RSRA | | Authors: | Zdanowski, K, Pecqueur, L, Werner, J, Potts, J.R, Kleanthous, C. | | Deposit date: | 2015-12-17 | | Release date: | 2016-08-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Anti-Sigma Factor Rsra Responds to Oxidative Stress by Reburying its Hydrophobic Core.
Nat.Commun., 7, 2016
|
|
2NSY
 
 | | CRYSTAL STRUCTURE OF NH3-DEPENDENT NAD+ SYNTHETASE FROM BACILLUS SUBTILIS IN COMPLEX WITH NAD-ADENYLATE | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Rizzi, M, Bolognesi, M, Coda, A. | | Deposit date: | 1998-07-14 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel deamido-NAD+-binding site revealed by the trapped NAD-adenylate intermediate in the NAD+ synthetase structure.
Structure, 6, 1998
|
|
1PKY
 
 | | PYRUVATE KINASE FROM E. COLI IN THE T-STATE | | Descriptor: | PYRUVATE KINASE | | Authors: | Mattevi, A. | | Deposit date: | 1995-04-27 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Escherichia coli pyruvate kinase type I: molecular basis of the allosteric transition.
Structure, 3, 1995
|
|
5FRF
 
 | | Solution structure of reduced and zinc-bound RsrA | | Descriptor: | ANTI-SIGMA FACTOR RSRA, ZINC ION | | Authors: | Zdanowski, K, Pecqueur, L, Werner, J, Potts, J.R, Kleanthous, C. | | Deposit date: | 2015-12-17 | | Release date: | 2016-08-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Anti-Sigma Factor Rsra Responds to Oxidative Stress by Reburying its Hydrophobic Core.
Nat.Commun., 7, 2016
|
|
8ROL
 
 | |
8RO9
 
 | | Human cohesin SMC1A-HD(longCC-EQ)/RAD21-C complex - Open/closed P-loop conformation | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 64-kDa C-terminal product, Structural maintenance of chromosomes protein 1A | | Authors: | Vitoria Gomes, M, Romier, C. | | Deposit date: | 2024-01-11 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.767 Å) | | Cite: | The cohesin ATPase cycle is mediated by specific conformational dynamics and interface plasticity of SMC1A and SMC3 ATPase domains.
Cell Rep, 43, 2024
|
|
8ROE
 
 | |
8ROA
 
 | |
