2N0N
 
 | | NMR solution structure for lactam (5,9) 11mer | | Descriptor: | lactam (5,9) 11mer peptide | | Authors: | Hoang, H.N, Song, K, Hill, T.A, Derksen, D.R, Edmonds, D.J, Kok, W.M, Limberakis, C, Liras, S, Loria, P.M, Mascitti, V, Mathiowetz, A.M, Mitchell, J.M, Piotrowski, D.W, Price, D.A, Stanton, R.V, Suen, J.Y, Withka, J.M, Griffith, D.A, Fairlie, D.P. | | Deposit date: | 2015-03-10 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Short Hydrophobic Peptides with Cyclic Constraints Are Potent Glucagon-like Peptide-1 Receptor (GLP-1R) Agonists.
J.Med.Chem., 58, 2015
|
|
2N08
 
 | | NMR structure of a short hydrophobic 11mer peptide in 25 mM SDS solution | | Descriptor: | Short hydrophobic peptide with cyclic constraints | | Authors: | Hoang, H.N, Song, K, Hill, T.A, Derksen, D.R, Edmonds, D.J, Kok, W.M, Limberakis, C, Liras, S, Loria, P.M, Mascitti, V, Mathiowetz, A.M, Mitchell, J.M, Piotrowski, D.W, Price, D.A, Stanton, R.V, Suen, J.Y, Withka, J.M, Griffith, D.A, Fairlie, D.P. | | Deposit date: | 2015-03-04 | | Release date: | 2015-04-15 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Short Hydrophobic Peptides with Cyclic Constraints Are Potent Glucagon-like Peptide-1 Receptor (GLP-1R) Agonists.
J.Med.Chem., 58, 2015
|
|
2N0I
 
 | | NMR solution structure for di-sulfide 11mer peptide | | Descriptor: | di-sulfide 11mer peptide | | Authors: | Hoang, H.N, Song, K, Hill, T.A, Derksen, D.R, Edmonds, D.J, Kok, W.M, Limberakis, C, Liras, S, Loria, P.M, Mascitti, V, Mathiowetz, A.M, Mitchell, J.M, Piotrowski, D.W, Price, D.A, Stanton, R.V, Suen, J.Y, Withka, J.M, Griffith, D.A, Fairlie, D.P. | | Deposit date: | 2015-03-09 | | Release date: | 2015-04-15 | | Last modified: | 2024-11-27 | | Method: | SOLUTION NMR | | Cite: | Short Hydrophobic Peptides with Cyclic Constraints Are Potent Glucagon-like Peptide-1 Receptor (GLP-1R) Agonists.
J.Med.Chem., 58, 2015
|
|
2N09
 
 | | NMR structure of a short hydrophobic 11mer peptide in DMSO-d6/H2O (1:3) solution | | Descriptor: | Short hydrophobic peptide with cyclic constraints | | Authors: | Hoang, H.N, Song, K, Hill, T.A, Derksen, D.R, Edmonds, D.J, Kok, W.M, Limberakis, C, Liras, S, Loria, P.M, Mascitti, V, Mathiowetz, A.M, Mitchell, J.M, Piotrowski, D.W, Price, D.A, Stanton, R.V, Suen, J.Y, Withka, J.M, Griffith, D.A, Fairlie, D.P. | | Deposit date: | 2015-03-04 | | Release date: | 2015-04-15 | | Last modified: | 2025-03-26 | | Method: | SOLUTION NMR | | Cite: | Short Hydrophobic Peptides with Cyclic Constraints Are Potent Glucagon-like Peptide-1 Receptor (GLP-1R) Agonists.
J.Med.Chem., 58, 2015
|
|
3O4L
 
 | | Genetic and structural basis for selection of a ubiquitous T cell receptor deployed in Epstein-Barr virus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BSLF2/BMLF1 protein, Beta-2-microglobulin, ... | | Authors: | Miles, J.J, Bulek, A.M, Cole, D.K, Gostick, E, Schauenburg, J.A, Dolton, G, Venturi, V, Davenport, M.P, Tan, M.P, Burrows, S.R, Wooldridge, L, Price, D.A, Rizkallah, P.J, Sewell, A.K. | | Deposit date: | 2010-07-27 | | Release date: | 2011-01-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Genetic and structural basis for selection of a ubiquitous T cell receptor deployed in Epstein-Barr virus infection.
Plos Pathog., 6, 2010
|
|
3RWH
 
 | | Rhesus macaque MHC class I molecule Mamu-B*17-MF8 | | Descriptor: | Beta-2-microglobulin, Major histocompatibility complex class I, Pol MF8 peptide from Pol protein | | Authors: | Wu, Y, Gao, F, Liu, J, Qi, J.X, Price, D.A, Gao, G.F. | | Deposit date: | 2011-05-09 | | Release date: | 2012-03-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of diverse peptide accommodation by the rhesus macaque MHC class I molecule Mamu-B*17: insights into immune protection from simian immunodeficiency virus
J.Immunol., 187, 2011
|
|
3RWG
 
 | | Rhesus macaque MHC class I molecule Mamu-B*17-MW9 | | Descriptor: | 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wu, Y, Gao, F, Liu, J, Qi, J.X, Price, D.A, Gao, G.F. | | Deposit date: | 2011-05-09 | | Release date: | 2012-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of diverse peptide accommodation by the rhesus macaque MHC class I molecule Mamu-B*17: insights into immune protection from simian immunodeficiency virus
J.Immunol., 187, 2011
|
|
3RWC
 
 | | Crystal structure of rhesus macaque MHC class I molecule Mamu-B*17-IW9 | | Descriptor: | Beta-2-microglobulin, Major histocompatibility complex class I, Nef IW9 peptide from Protein Nef | | Authors: | Wu, Y, Gao, F, Liu, J, Qi, J.X, Price, D.A, Gao, G.F. | | Deposit date: | 2011-05-09 | | Release date: | 2012-03-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structural basis of diverse peptide accommodation by the rhesus macaque MHC class I molecule Mamu-B*17: insights into immune protection from simian immunodeficiency virus
J.Immunol., 187, 2011
|
|
3RWI
 
 | | Rhesus macaque MHC class I molecule Mamu-B*17-GW10 | | Descriptor: | Beta-2-microglobulin, Major histocompatibility complex class I, Vif GW10 peptide from Virion infectivity factor | | Authors: | Wu, Y, Gao, F, Liu, J, Qi, J.X, Price, D.A, Gao, G.F. | | Deposit date: | 2011-05-09 | | Release date: | 2012-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.009 Å) | | Cite: | Structural basis of diverse peptide accommodation by the rhesus macaque MHC class I molecule Mamu-B*17: insights into immune protection from simian immunodeficiency virus
J.Immunol., 187, 2011
|
|
3RWJ
 
 | | Rhesus macaque MHC class I molecule Mamu-B*17-HW8 | | Descriptor: | Beta-2-microglobulin, Major histocompatibility complex class I, Vif HW8 peptide from Virion infectivity factor | | Authors: | Wu, Y, Gao, F, Liu, J, Qi, J.X, Price, D.A, Gao, G.F. | | Deposit date: | 2011-05-09 | | Release date: | 2012-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of diverse peptide accommodation by the rhesus macaque MHC class I molecule Mamu-B*17: insights into immune protection from simian immunodeficiency virus
J.Immunol., 187, 2011
|
|
3RWE
 
 | | rhesus macaque MHC class I molecule Mamu-B*17-FW9 | | Descriptor: | Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, Major histocompatibility complex class I, ... | | Authors: | Wu, Y, Gao, F, Liu, J, Qi, J.X, Price, D.A, Gao, G.F. | | Deposit date: | 2011-05-09 | | Release date: | 2012-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of diverse peptide accommodation by the rhesus macaque MHC class I molecule Mamu-B*17: insights into immune protection from simian immunodeficiency virus
J.Immunol., 187, 2011
|
|
3RWD
 
 | | rhesus macaque MHC class I molecule Mamu-B*17-IW11 | | Descriptor: | Beta-2-microglobulin, Major histocompatibility complex class I, Nef IW11 peptide from Protein Nef | | Authors: | Wu, Y, Gao, F, Liu, J, Qi, J.X, Price, D.A, Gao, G.F. | | Deposit date: | 2011-05-09 | | Release date: | 2012-03-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Structural basis of diverse peptide accommodation by the rhesus macaque MHC class I molecule Mamu-B*17: insights into immune protection from simian immunodeficiency virus
J.Immunol., 187, 2011
|
|
3RWF
 
 | | Rhesus macaque MHC class I molecule Mamu-B*17-QW9 | | Descriptor: | Beta-2-microglobulin, Major histocompatibility complex class I, Nef QW9 peptide from Protein Nef | | Authors: | Wu, Y, Gao, F, Liu, J, Qi, J.X, Price, D.A, Gao, G.F. | | Deposit date: | 2011-05-09 | | Release date: | 2012-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of diverse peptide accommodation by the rhesus macaque MHC class I molecule Mamu-B*17: insights into immune protection from simian immunodeficiency virus
J.Immunol., 187, 2011
|
|
3HG1
 
 | | Germline-governed recognition of a cancer epitope by an immunodominant human T cell receptor | | Descriptor: | Beta-2-microglobulin, CANCER/MART-1, GLYCEROL, ... | | Authors: | Cole, D.K, Yuan, F, Rizkallah, P.J, Miles, J.J, Gostick, E, Price, D.A, Gao, G.F, Jakobsen, B.K, Sewell, A.K. | | Deposit date: | 2009-05-13 | | Release date: | 2009-07-28 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Germ line-governed recognition of a cancer epitope by an immunodominant human T-cell receptor.
J.Biol.Chem., 284, 2009
|
|
4GKS
 
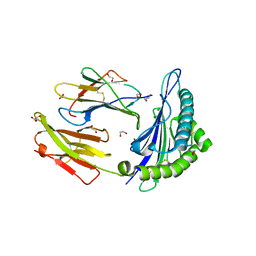 | | A2-MHC Complex carrying FLTGIGIITV | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, FLT Cognate peptide, ... | | Authors: | Sewell, A.K, Rizkallah, P.J, Cole, D.K, Wooldridge, L, Price, D.A. | | Deposit date: | 2012-08-13 | | Release date: | 2012-09-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.346 Å) | | Cite: | T-cell Receptor-optimized Peptide Skewing of the T-cell Repertoire Can Enhance Antigen Targeting.
J.Biol.Chem., 287, 2012
|
|
4GKN
 
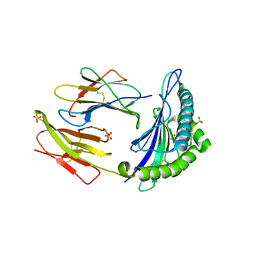 | | A2-MHC Complex carrying FATGIGIITV | | Descriptor: | Beta-2-microglobulin, FAT Cognate peptide, MHC class I antigen, ... | | Authors: | Sewell, A.K, Rizkallah, P.J, Cole, D.K, Wooldridge, L, Price, D.A. | | Deposit date: | 2012-08-13 | | Release date: | 2012-09-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.753 Å) | | Cite: | T-cell Receptor-optimized Peptide Skewing of the T-cell Repertoire Can Enhance Antigen Targeting.
J.Biol.Chem., 287, 2012
|
|
6VQD
 
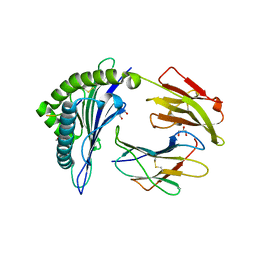 | | HLA-B*27:05 presenting an HIV-1 8mer peptide | | Descriptor: | 8-mer peptide, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Pymm, P, Tenzer, S, Wee, E, Weimershaus, M, Burgevin, A, Kollnberger, S, Gerstoft, J, Josephs, T.M, Ladell, K, Mclaren, J.E, Appay, V, Price, D.A, Fugger, L, Bell, J.I, Hansjorg, S, Van Endert, P, Harkiolaki, M, Iversen, A.K.N. | | Deposit date: | 2020-02-05 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Epitope length variants balance protective immune responses and viral escape in HIV-1 infection
Cell Rep, 38, 2022
|
|
6VQY
 
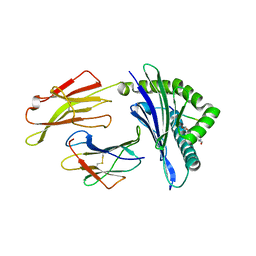 | | HLA-B*27:05 presenting an HIV-1 7mer peptide | | Descriptor: | 7-mer peptide, ARGININE, Beta-2-microglobulin, ... | | Authors: | Pymm, P, Tenzer, S, Wee, E, Weimershaus, M, Burgevin, A, Kollnberger, S, Gerstoft, J, Josephs, T.M, Ladell, K, Mclaren, J.E, Appay, V, Price, D.A, Fugger, L, Bell, J.I, Hansjorg, S, Van Endert, P, Harkiolaki, M, Iversen, A.K.N. | | Deposit date: | 2020-02-06 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Epitope length variants balance protective immune responses and viral escape in HIV-1 infection
Cell Rep, 38, 2022
|
|
6VQ2
 
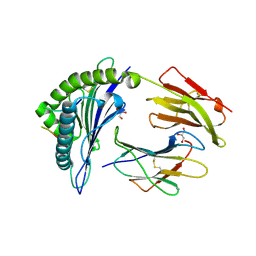 | | HLA-B*27:05 presenting an HIV-1 14mer peptide | | Descriptor: | 14-mer peptide, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Pymm, P, Tenzer, S, Wee, E, Weimershaus, M, Burgevin, A, Kollnberger, S, Gerstoft, J, Josephs, T.M, Ladell, K, Mclaren, J.E, Appay, V, Price, D.A, Fugger, L, Bell, J.I, Hansjorg, S, Van Endert, P, Harkiolaki, M, Iversen, A.K.N. | | Deposit date: | 2020-02-04 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Epitope length variants balance protective immune responses and viral escape in HIV-1 infection
Cell Rep, 38, 2022
|
|
6VQE
 
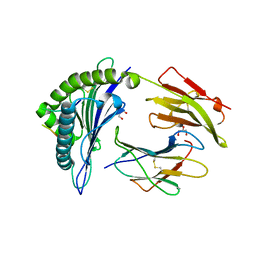 | | HLA-B*27:05 presenting an HIV-1 13mer peptide | | Descriptor: | 13-mer peptide, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Pymm, P, Tenzer, S, Wee, E, Weimershaus, M, Burgevin, A, Kollnberger, S, Gerstoft, J, Josephs, T.M, Ladell, K, Mclaren, J.E, Appay, V, Price, D.A, Fugger, L, Bell, J.I, Hansjorg, S, Van Endert, P, Harkiolaki, M, Iversen, A.K.N. | | Deposit date: | 2020-02-05 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Epitope length variants balance protective immune responses and viral escape in HIV-1 infection
Cell Rep, 38, 2022
|
|
6VQZ
 
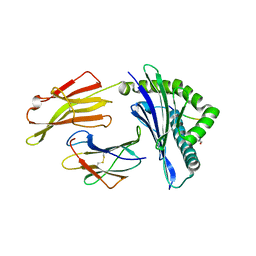 | | HLA-B*27:05 presenting an HIV-1 6mer peptide | | Descriptor: | 6-mer peptide, ARGININE, Beta-2-microglobulin, ... | | Authors: | Pymm, P, Tenzer, S, Wee, E, Weimershaus, M, Burgevin, A, Kollnberger, S, Gerstoft, J, Josephs, T.M, Ladell, K, Mclaren, J.E, Appay, V, Price, D.A, Fugger, L, Bell, J.I, Hansjorg, S, Van Endert, P, Harkiolaki, M, Iversen, A.K.N. | | Deposit date: | 2020-02-06 | | Release date: | 2021-02-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Epitope length variants balance protective immune responses and viral escape in HIV-1 infection
Cell Rep, 38, 2022
|
|
6VPZ
 
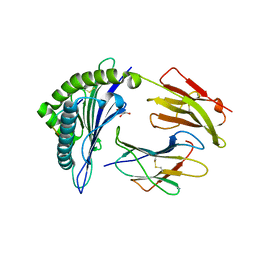 | | HLA-B*27:05 presenting an HIV-1 11mer peptide | | Descriptor: | 11-mer peptide, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Pymm, P, Tenzer, S, Wee, E, Weimershaus, M, Burgevin, A, Kollnberger, S, Gerstoft, J, Josephs, T.M, Ladell, K, Mclaren, J.E, Appay, V, Price, D.A, Fugger, L, Bell, J.I, Hansjorg, S, Van Endert, P, Harkiolaki, M, Iversen, A.K.N. | | Deposit date: | 2020-02-04 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Epitope length variants balance protective immune responses and viral escape in HIV-1 infection
Cell Rep, 38, 2022
|
|
5DI1
 
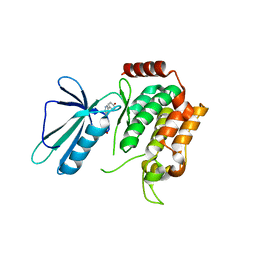 | | MAP4K4 in complex with an inhibitor | | Descriptor: | 4-{6-amino-5-[4-(methylsulfonyl)phenyl]pyridin-3-yl}phenol, Mitogen-activated protein kinase kinase kinase kinase 4 | | Authors: | Liu, S. | | Deposit date: | 2015-08-31 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of an in Vivo Tool to Establish Proof-of-Concept for MAP4K4-Based Antidiabetic Treatment.
Acs Med.Chem.Lett., 6, 2015
|
|
5JQ1
 
 | |
5JPV
 
 | |
