1B7D
 
 | | NEUROTOXIN (TS1) FROM BRAZILIAN SCORPION TITYUS SERRULATUS | | Descriptor: | PHOSPHATE ION, PROTEIN (NEUROTOXIN TS1) | | Authors: | Polikarpov, I, Sanches Jr, M.S, Marangoni, S, Toyama, M.H, Teplyakov, A. | | Deposit date: | 1999-01-21 | | Release date: | 1999-07-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of neurotoxin Ts1 from Tityus serrulatus provides insights into the specificity and toxicity of scorpion toxins.
J.Mol.Biol., 290, 1999
|
|
8GF3
 
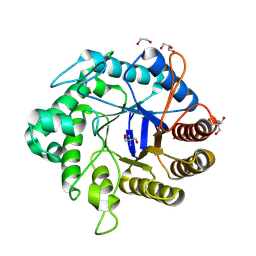 | | Crystallographic structure from BlMan5_7 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GH5 Mannanase, ... | | Authors: | Briganti, L, Araujo, E.A, Polikarpov, I. | | Deposit date: | 2023-03-07 | | Release date: | 2024-04-17 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Unravelling biochemical and structural features of Bacillus licheniformis GH5 mannanase using site-directed mutagenesis and high-resolution protein crystallography studies.
Int.J.Biol.Macromol., 274, 2024
|
|
4CI4
 
 | |
1QLR
 
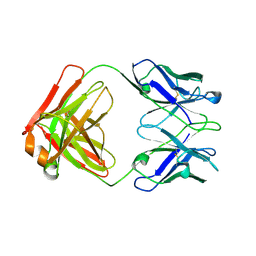 | | CRYSTAL STRUCTURE OF THE FAB FRAGMENT OF A HUMAN MONOCLONAL IgM COLD AGGLUTININ | | Descriptor: | IGM FAB REGION IV-J(H4)-C (KAU COLD AGGLUTININ), IGM KAPPA CHAIN V-III (KAU COLD AGGLUTININ), alpha-L-fucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Carvalho, J.G, Cauerhff, A, Goldbaum, F, Leoni, J, Polikarpov, I. | | Deposit date: | 1999-09-11 | | Release date: | 2000-09-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Three-dimensional structure of the Fab from a human IgM cold agglutinin.
J Immunol., 165, 2000
|
|
4USC
 
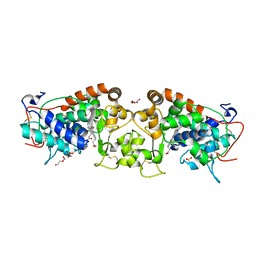 | | Crystal structure of peroxidase from palm tree Chamaerops excelsa | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Bernardes, A, Santos, J.C, Textor, L.C, Cuadrado, N.H, Kostetsky, E.Y, Roig, M.G, Muniz, J.R.C, Shnyrov, V.L, Polikarpov, I. | | Deposit date: | 2014-07-07 | | Release date: | 2015-05-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure Analysis of Peroxidase from the Palm Tree Chamaerops Excelsa.
Biochimie, 111, 2015
|
|
8UWS
 
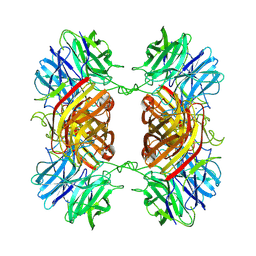 | | Cryo-EM structure of an Enterobacter GH43 Beta-Xylosidase: EcXyl43 | | Descriptor: | Beta-xylosidase, CALCIUM ION | | Authors: | Briganti, L, Godoy, A.S, Capetti, C.C.M, Portugal, R.V, Polikarpov, I. | | Deposit date: | 2023-11-08 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Cryo-EM structure of an Enterobacter GH43 Beta-Xylosidase: EcXyl43
To Be Published
|
|
4LNW
 
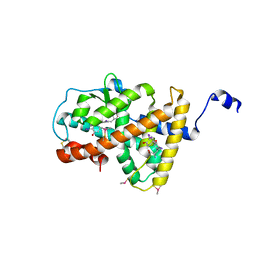 | | Crystal structure of TR-alpha bound to T3 in a second site | | Descriptor: | 3,5,3'TRIIODOTHYRONINE, Thyroid hormone receptor alpha | | Authors: | Puhl, A.C, Aparicio, R, Polikarpov, I. | | Deposit date: | 2013-07-12 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of a new hormone-binding site on the surface of thyroid hormone receptor.
Mol.Endocrinol., 28, 2014
|
|
4LNX
 
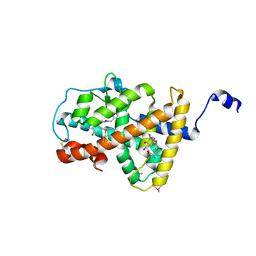 | | Crystal structure of TR-alpha bound to T4 in a second site | | Descriptor: | 3,5,3',5'-TETRAIODO-L-THYRONINE, 3,5,3'TRIIODOTHYRONINE, Thyroid hormone receptor alpha | | Authors: | Puhl, A.C, Aparicio, R, Polikarpov, I. | | Deposit date: | 2013-07-12 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Identification of a new hormone-binding site on the surface of thyroid hormone receptor.
Mol.Endocrinol., 28, 2014
|
|
4UZS
 
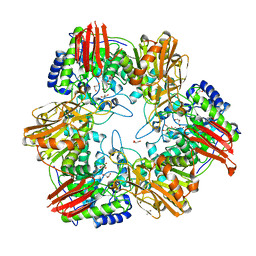 | | Crystal structure of Bifidobacterium bifidum beta-galactosidase | | Descriptor: | BETA-GALACTOSIDASE, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Godoy, A.S, Murakami, M.T, Camilo, C.M, Bernardes, A, Polikarpov, I. | | Deposit date: | 2014-09-08 | | Release date: | 2015-09-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal Structure of Beta1-6-Galactosidase from Bifidobacterium Bifidum S17: Trimeric Architecture, Molecular Determinants of the Enzymatic Activity and its Inhibition by Alpah-Galactose.
FEBS J., 283, 2016
|
|
3KGU
 
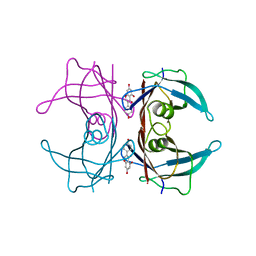 | |
3KGT
 
 | |
3KGS
 
 | | V30M mutant human transthyretin (TTR) (apoV30M) pH 7.5 | | Descriptor: | Transthyretin | | Authors: | Trivella, D.B, Polikarpov, I. | | Deposit date: | 2009-10-29 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformational differences between the wild type and V30M mutant transthyretin modulate its binding to genistein: implications to tetramer stability and ligand-binding.
J.Struct.Biol., 170, 2010
|
|
6MVI
 
 | | Apo Cel45A from Neurospora crassa OR74A | | Descriptor: | Endoglucanase V | | Authors: | Kadowaki, M.A.S, Polikarpov, I. | | Deposit date: | 2018-10-25 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural insights into the hydrolysis pattern and molecular dynamics simulations of GH45 subfamily a endoglucanase from Neurospora crassa OR74A.
Biochimie, 165, 2019
|
|
6BUS
 
 | |
4XTA
 
 | |
6MVJ
 
 | | Cellobiose complex Cel45A from Neurospora crassa OR74A | | Descriptor: | Endoglucanase V, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Kadowaki, M.A.S, Polikarpov, I. | | Deposit date: | 2018-10-25 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.809 Å) | | Cite: | Structural insights into the hydrolysis pattern and molecular dynamics simulations of GH45 subfamily a endoglucanase from Neurospora crassa OR74A.
Biochimie, 165, 2019
|
|
4XUM
 
 | |
6O9N
 
 | | Structural insights on a new fungal aryl-alcohol oxidase | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kadowaki, M.A.S, Polikarpov, I. | | Deposit date: | 2019-03-14 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Enzymatic versatility and thermostability of a new aryl-alcohol oxidase from Thermothelomyces thermophilus M77.
Biochim Biophys Acta Gen Subj, 1864, 2020
|
|
4OJ4
 
 | |
1QFE
 
 | | THE STRUCTURE OF TYPE I 3-DEHYDROQUINATE DEHYDRATASE FROM SALMONELLA TYPHI | | Descriptor: | 3-AMINO-4,5-DIHYDROXY-CYCLOHEX-1-ENECARBOXYLATE, PROTEIN (3-DEHYDROQUINATE DEHYDRATASE) | | Authors: | Shrive, A.K, Polikarpov, I, Sawyer, L, Coggins, J.R, Hawkins, A.R. | | Deposit date: | 1999-04-05 | | Release date: | 2000-04-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The two types of 3-dehydroquinase have distinct structures but catalyze the same overall reaction.
Nat.Struct.Biol., 6, 1999
|
|
1QG5
 
 | |
1QLL
 
 | | Piratoxin-II (Prtx-II) - a K49 PLA2 from Bothrops pirajai | | Descriptor: | N-TRIDECANOIC ACID, PHOSPHOLIPASE A2 | | Authors: | Lee, W.-H, Polikarpov, I. | | Deposit date: | 1999-09-01 | | Release date: | 2000-09-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Basis for Low Catalytic Activity in Lys49 Phospholipases A2-A Hypothesis: The Crystal Structure of Piratoxin II Complexed to Fatty Acid
Biochemistry, 40, 2001
|
|
8U4F
 
 | |
8U4A
 
 | |
8U49
 
 | | The Apo Crystal Structure of BlCel9A from Glycoside Hydrolase Family 9 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Araujo, E.A, Polikarpov, I. | | Deposit date: | 2023-09-10 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular mechanism of cellulose depolymerization by the two-domain BlCel9A enzyme from the glycoside hydrolase family 9.
Carbohydr Polym, 329, 2024
|
|
