1A37
 
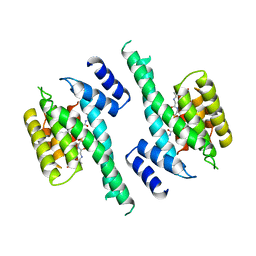 | | 14-3-3 PROTEIN ZETA BOUND TO PS-RAF259 PEPTIDE | | Descriptor: | 14-3-3 PROTEIN ZETA, PS-RAF259 PEPTIDE LSQRQRST(SEP)TPNVHM | | Authors: | Petosa, C, Masters, S.C, Pohl, J, Wang, B, Fu, H, Liddington, R.C. | | Deposit date: | 1998-01-28 | | Release date: | 1999-03-02 | | Last modified: | 2016-08-17 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | 14-3-3zeta binds a phosphorylated Raf peptide and an unphosphorylated peptide via its conserved amphipathic groove.
J.Biol.Chem., 273, 1998
|
|
1A38
 
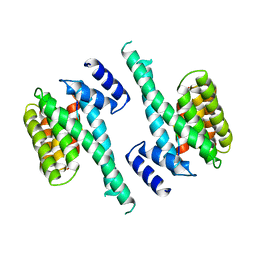 | | 14-3-3 PROTEIN ZETA BOUND TO R18 PEPTIDE | | Descriptor: | 14-3-3 PROTEIN ZETA, R18 PEPTIDE (PHCVPRDLSWLDLEANMCLP) | | Authors: | Petosa, C, Masters, S.C, Pohl, J, Wang, B, Fu, H, Liddington, R.C. | | Deposit date: | 1998-01-28 | | Release date: | 1999-03-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | 14-3-3zeta binds a phosphorylated Raf peptide and an unphosphorylated peptide via its conserved amphipathic groove.
J.Biol.Chem., 273, 1998
|
|
1ACC
 
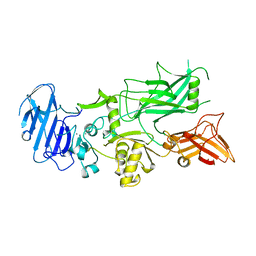 | | ANTHRAX PROTECTIVE ANTIGEN | | Descriptor: | ANTHRAX PROTECTIVE ANTIGEN, CALCIUM ION | | Authors: | Petosa, C, Liddington, R.C. | | Deposit date: | 1997-02-05 | | Release date: | 1998-02-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the anthrax toxin protective antigen.
Nature, 385, 1997
|
|
2C9L
 
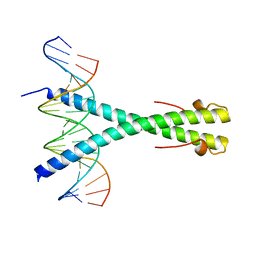 | | Structure of the Epstein-Barr virus ZEBRA protein | | Descriptor: | 5'-D(*AP*AP*GP*CP*AP*CP*TP*GP*AP*CP *TP*CP*AP*TP*GP*AP*AP*GP*T)-3', 5'-D(*AP*CP*TP*TP*CP*AP*CP*TP*GP*AP *GP*TP*CP*AP*GP*TP*GP*CP*T)-3', BZLF1 TRANS-ACTIVATOR PROTEIN | | Authors: | Petosa, C, Morand, P, Baudin, F, Moulin, M, Artero, J.B, Muller, C.W. | | Deposit date: | 2005-12-13 | | Release date: | 2006-02-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Basis of Lytic Cycle Activation by the Epstein-Barr Virus Zebra Protein
Mol.Cell, 21, 2006
|
|
2C9N
 
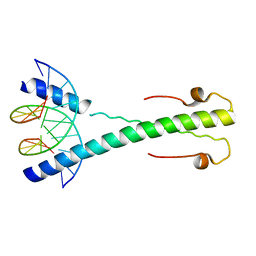 | | Structure of the Epstein-Barr virus ZEBRA protein at approximately 3. 5 Angstrom resolution | | Descriptor: | 5'-D(*CP*AP*CP*TP*GP*AP*CP*TP*CP*AP *T)-3', 5'-D(*CP*AP*TP*GP*AP*GP*TP*CP*AP*GP *T)-3', BZLF1 TRANS-ACTIVATOR PROTEIN | | Authors: | Petosa, C, Morand, P, Baudin, F, Moulin, M, Artero, J.B, Muller, C.W. | | Deposit date: | 2005-12-13 | | Release date: | 2006-02-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Basis of Lytic Cycle Activation by the Epstein-Barr Virus Zebra Protein
Mol.Cell, 21, 2006
|
|
1W9C
 
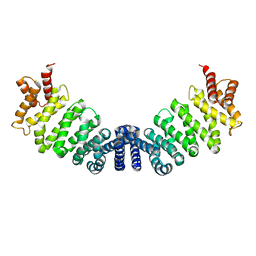 | | Proteolytic fragment of CRM1 spanning six C-terminal HEAT repeats | | Descriptor: | CRM1 PROTEIN | | Authors: | Petosa, C, Schoehn, G, Askjaer, P, Bauer, U, Moulin, M, Steuerwald, U, Soler-Lopez, M, Baudin, F, Mattaj, I.W, Muller, C.W. | | Deposit date: | 2004-10-08 | | Release date: | 2004-12-03 | | Last modified: | 2019-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Architecture of Crm1-Exportin 1 Suggests How Cooperativity is Achieved During Formation of a Nuclear Export Complex
Mol.Cell, 16, 2004
|
|
7NX5
 
 | |
2PYO
 
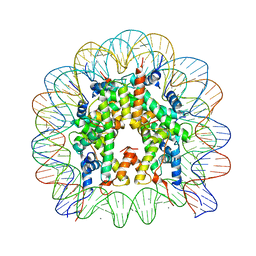 | | Drosophila nucleosome core | | Descriptor: | CHLORIDE ION, DNA (147-MER), Histone H2A, ... | | Authors: | Clapier, C.R, Petosa, C, Mueller, C.W. | | Deposit date: | 2007-05-16 | | Release date: | 2007-11-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structure of the Drosophila nucleosome core particle highlights evolutionary constraints on the H2A-H2B histone dimer.
Proteins, 71, 2007
|
|
7ZL8
 
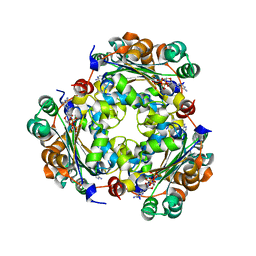 | | NME1 in complex with succinyl-CoA | | Descriptor: | Nucleoside diphosphate kinase A, SUCCINYL-COENZYME A | | Authors: | Garcia-Saez, I, Iuso, D, Khochbin, S, Petosa, C. | | Deposit date: | 2022-04-14 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Nucleoside diphosphate kinases 1 and 2 regulate a protective liver response to a high-fat diet.
Sci Adv, 9, 2023
|
|
7ZLW
 
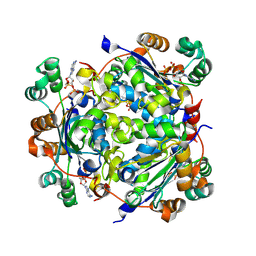 | | NME1 in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Nucleoside diphosphate kinase A | | Authors: | Garcia-Saez, I, Iuso, D, Khochbin, S, Petosa, C. | | Deposit date: | 2022-04-15 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Nucleoside diphosphate kinases 1 and 2 regulate a protective liver response to a high-fat diet.
Sci Adv, 9, 2023
|
|
7ZTK
 
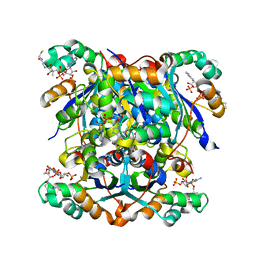 | | NME1 in complex with CoA | | Descriptor: | COENZYME A, Nucleoside diphosphate kinase A | | Authors: | Garcia-Saez, I, Iuso, D, Khochbin, S, Petosa, C. | | Deposit date: | 2022-05-10 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Nucleoside diphosphate kinases 1 and 2 regulate a protective liver response to a high-fat diet.
Sci Adv, 9, 2023
|
|
5NL0
 
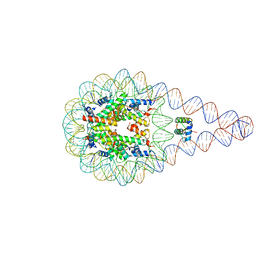 | | Crystal structure of a 197-bp palindromic 601L nucleosome in complex with linker histone H1 | | Descriptor: | DNA (197-MER), Histone H1.0-B, Histone H2A type 1, ... | | Authors: | Garcia-Saez, I, Petosa, C, Dimitrov, S. | | Deposit date: | 2017-04-03 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (5.4 Å) | | Cite: | Structure and Dynamics of a 197 bp Nucleosome in Complex with Linker Histone H1.
Mol. Cell, 66, 2017
|
|
6TEM
 
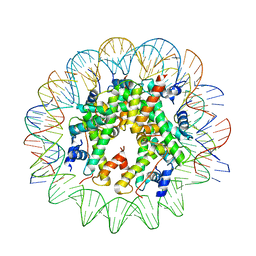 | | CENP-A nucleosome core particle with 145 base pairs of the Widom 601 sequence by cryo-EM | | Descriptor: | Histone H2A, Histone H2B 1.1, Histone H3-like centromeric protein A, ... | | Authors: | Boopathi, R, Danev, R, Petosa, C, Bednar, J. | | Deposit date: | 2019-11-12 | | Release date: | 2020-04-08 | | Last modified: | 2020-06-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Phase-plate cryo-EM structure of the Widom 601 CENP-A nucleosome core particle reveals differential flexibility of the DNA ends.
Nucleic Acids Res., 48, 2020
|
|
1A4O
 
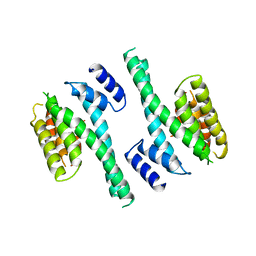 | | 14-3-3 PROTEIN ZETA ISOFORM | | Descriptor: | 14-3-3 PROTEIN ZETA | | Authors: | Liu, D, Bienkowska, J, Petosa, C, Collier, R.J, Fu, H, Liddington, R.C. | | Deposit date: | 1998-02-01 | | Release date: | 1999-03-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the zeta isoform of the 14-3-3 protein.
Nature, 376, 1995
|
|
6HKT
 
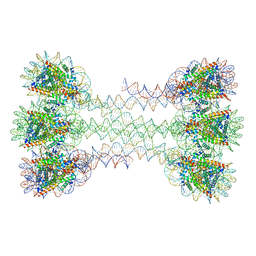 | | Structure of an H1-bound 6-nucleosome array | | Descriptor: | DNA (1122-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Garcia-Saez, I, Dimitrov, S, Petosa, C. | | Deposit date: | 2018-09-08 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (9.7 Å) | | Cite: | Structure of an H1-Bound 6-Nucleosome Array Reveals an Untwisted Two-Start Chromatin Fiber Conformation.
Mol. Cell, 72, 2018
|
|
4BSN
 
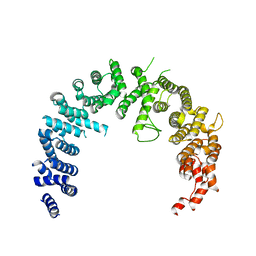 | | Crystal structure of the Nuclear Export Receptor CRM1 (exportin-1) lacking the C-terminal helical extension at 4.1A | | Descriptor: | EXPORTIN-1 | | Authors: | Dian, C, Bernaudat, F, Langer, K, Oliva, M.F, Fornerod, M, Schoehn, G, Muller, C.W, Petosa, C. | | Deposit date: | 2013-06-11 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structure of a Truncation Mutant of the Nuclear Export Factor Crm1 Provides Insights Into the Auto-Inhibitory Role of its C-Terminal Helix.
Structure, 21, 2013
|
|
4BSM
 
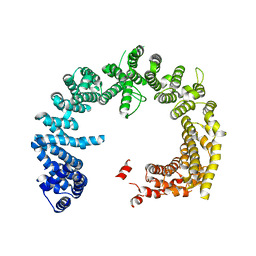 | | Crystal structure of the Nuclear Export Receptor CRM1 (exportin-1) lacking the C-terminal helical extension at 4.5A | | Descriptor: | EXPORTIN-1 | | Authors: | Dian, C, Bernaudat, F, Langer, K, Oliva, M.F, Fornerod, M, Schoehn, G, Muller, C.W, Petosa, C. | | Deposit date: | 2013-06-10 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Structure of a Truncation Mutant of the Nuclear Export Factor Crm1 Provides Insights Into the Auto-Inhibitory Role of its C-Terminal Helix.
Structure, 21, 2013
|
|
5N16
 
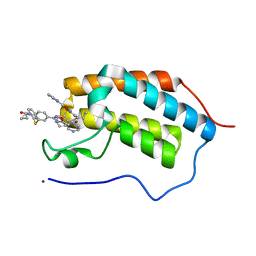 | | First Bromodomain (BD1) from Candida albicans Bdf1 bound to a dibenzothiazepinone (compound 1) | | Descriptor: | 5-cyclopropyl-2-(5-pyrazin-2-yl-1,2,4-oxadiazol-3-yl)benzo[b][1,4]benzothiazepin-6-one, Bromodomain-containing factor 1, GLYCEROL, ... | | Authors: | Mietton, F, Ferri, E, Champleboux, M, Zala, N, Maubon, D, Zhou, Y, Harbut, M, Spittler, D, Garnaud, C, Courcon, M, Chauvel, M, d'Enfert, C, Kashemirov, B.A, Hull, M, Cornet, M, McKenna, C.E, Govin, J, Petosa, C. | | Deposit date: | 2017-02-05 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Selective BET bromodomain inhibition as an antifungal therapeutic strategy.
Nat Commun, 8, 2017
|
|
5N13
 
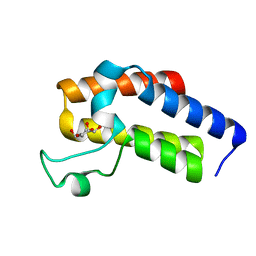 | | Second Bromodomain (BD2) from Candida albicans Bdf1 in the unbound form | | Descriptor: | Bromodomain-containing factor 1, GLYCEROL | | Authors: | Mietton, F, Ferri, E, Champlebouxm, M, Zala, N, Maubon, D, Zhou, Y, Harbut, M, Spittler, D, Garnaud, C, Courcon, M, Chauvel, M, d'Enfert, C, Kashemirov, B.A, Hull, M, Cornet, M, McKenna, C.E, Govin, J, Petosa, C. | | Deposit date: | 2017-02-04 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Selective BET bromodomain inhibition as an antifungal therapeutic strategy.
Nat Commun, 8, 2017
|
|
5N17
 
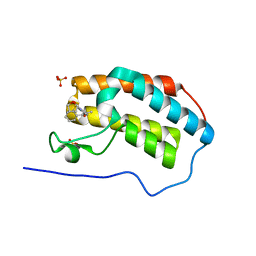 | | First Bromodomain (BD1) from Candida albicans Bdf1 bound to a dibenzothiazepinone (compound 3) | | Descriptor: | (2~{S})-~{N}-(5-methyl-6-oxidanylidene-benzo[b][1,4]benzothiazepin-2-yl)oxolane-2-carboxamide, Bromodomain-containing factor 1, SULFATE ION | | Authors: | Mietton, F, Ferri, E, Champleboux, M, Zala, N, Maubon, D, Zhou, Y, Harbut, M, Spittler, D, Garnaud, C, Courcon, M, Chauvon, M, d'Enfer, C, Kashemirov, B.A, Hull, M, Cornet, M, McKenna, C.E, Govin, J, Petosa, C. | | Deposit date: | 2017-02-05 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Selective BET bromodomain inhibition as an antifungal therapeutic strategy.
Nat Commun, 8, 2017
|
|
5N15
 
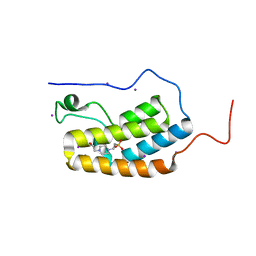 | | First Bromodomain (BD1) from Candida albicans Bdf1 in the unbound form | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Bromodomain-containing factor 1, GLYCEROL, ... | | Authors: | Mietton, F, Ferri, E, Champleboux, M, Zala, N, Maubon, D, Zhou, Y, Harbut, M, Spittler, D, Garnaud, C, Courcon, M, Chauvel, M, d'Enfert, C, Kashemirov, B.A, Hull, M, Cornet, M, McKenna, C.E, Govin, J, Petosa, C. | | Deposit date: | 2017-02-05 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Selective BET bromodomain inhibition as an antifungal therapeutic strategy.
Nat Commun, 8, 2017
|
|
5N18
 
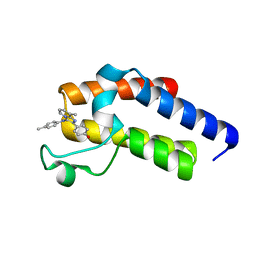 | | Second Bromodomain (BD2) from Candida albicans Bdf1 bound to an imidazopyridine (compound 2) | | Descriptor: | 4-[8-methyl-3-[(4-methylphenyl)amino]imidazo[1,2-a]pyridin-2-yl]phenol, Bromodomain-containing factor 1, GLYCEROL | | Authors: | Mietton, F, Ferri, E, Champleboux, M, Zala, N, Maubon, D, Zhou, Y, Harbut, M, Spittler, D, Garnaud, C, Courcon, M, Chauvel, M, d'Enfert, C, Kashemirov, B.A, Hull, M, Cornet, M, McKenna, C.E, Govin, J, Petosa, C. | | Deposit date: | 2017-02-05 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Selective BET bromodomain inhibition as an antifungal therapeutic strategy.
Nat Commun, 8, 2017
|
|
2Y8F
 
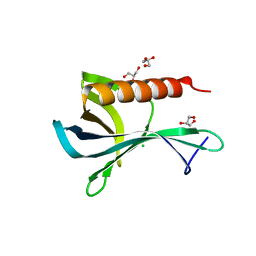 | | Structure of the Ran-binding domain from human RanBP3 (wild type) | | Descriptor: | CHLORIDE ION, GLYCEROL, RAN-BINDING PROTEIN 3 | | Authors: | Langer, K, Dian, C, Rybin, V, Muller, C.W, Petosa, C. | | Deposit date: | 2011-02-06 | | Release date: | 2011-02-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights Into the Function of the Crm1 Cofactor Ranbp3 from the Structure of its Ran-Binding Domain
Plos One, 6, 2011
|
|
2Y8G
 
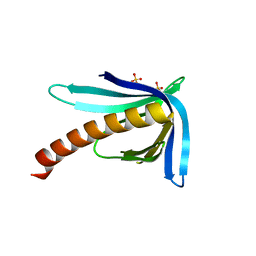 | | Structure of the Ran-binding domain from human RanBP3 (E352A-R353V double mutant) | | Descriptor: | RAN-BINDING PROTEIN 3, SULFATE ION | | Authors: | Langer, K, Dian, C, Rybin, V, Muller, C.W, Petosa, C. | | Deposit date: | 2011-02-06 | | Release date: | 2011-02-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Insights Into the Function of the Crm1 Cofactor Ranbp3 from the Structure of its Ran-Binding Domain
Plos One, 6, 2011
|
|
1UUS
 
 | | Structure of an activated Dictyostelium STAT in its DNA-unbound form | | Descriptor: | STAT PROTEIN | | Authors: | Soler-Lopez, M, Petosa, C, Fukuzawa, M, Ravelli, R, Williams, J.G, Muller, C.W. | | Deposit date: | 2004-01-09 | | Release date: | 2004-03-26 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of an Activated Dictyostelium Stat in its DNA-Unbound Form
Mol.Cell, 13, 2004
|
|
