1XQI
 
 | | Crystal Structure Analysis of an NDP kinase from Pyrobaculum aerophilum | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Nucleoside diphosphate kinase, TRIETHYLENE GLYCOL | | Authors: | Pedelacq, J.D, Waldo, G.S, Cabantous, S, Liong, E.C, Berendzen, J, Terwilliger, T.C. | | Deposit date: | 2004-10-12 | | Release date: | 2005-09-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional features of an NDP kinase from the hyperthermophile crenarchaeon Pyrobaculum aerophilum
Protein Sci., 14, 2005
|
|
2B3P
 
 | | Crystal structure of a superfolder green fluorescent protein | | Descriptor: | ACETIC ACID, CADMIUM ION, green fluorescent protein | | Authors: | Pedelacq, J.D, Cabantous, S, Tran, T.H, Terwilliger, T.C, Waldo, G.S. | | Deposit date: | 2005-09-20 | | Release date: | 2005-11-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Engineering and characterization of a superfolder green fluorescent protein.
Nat.Biotechnol., 24, 2006
|
|
2B3Q
 
 | | Crystal structure of a well-folded variant of green fluorescent protein | | Descriptor: | MAGNESIUM ION, green fluorescent protein | | Authors: | Pedelacq, J.D, Cabantous, S, Tran, T.H, Terwilliger, T.C, Waldo, G.S. | | Deposit date: | 2005-09-20 | | Release date: | 2005-11-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Engineering and characterization of a superfolder green fluorescent protein.
Nat.Biotechnol., 24, 2006
|
|
1PVL
 
 | | STRUCTURE OF THE PANTON-VALENTINE LEUCOCIDIN F COMPONENT FROM STAPHYLOCOCCUS AUREUS | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, LEUCOCIDIN | | Authors: | Pedelacq, J.D, Mourey, L, Maveyraud, L, Prevost, G, Samama, J.P. | | Deposit date: | 1999-01-12 | | Release date: | 1999-06-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of a Staphylococcus aureus leucocidin component (LukF-PV) reveals the fold of the water-soluble species of a family of transmembrane pore-forming toxins.
Structure Fold.Des., 7, 1999
|
|
4U89
 
 | |
8QZJ
 
 | |
8QZH
 
 | | Crystal structure of apo-PptT from Mycobacterium tuberculosis | | Descriptor: | 4'-phosphopantetheinyl transferase PptT, ACETATE ION, MAGNESIUM ION | | Authors: | Gavalda, S, Carivenc, C, Mourey, L, Pedelacq, J.D. | | Deposit date: | 2023-10-27 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Catalytic Cycle of Type II 4'-Phosphopantetheinyl Transferases
Acs Catalysis, 14, 2024
|
|
8QZI
 
 | | Crystal structure of PptT-ACP from Mycobacterium tuberculosis | | Descriptor: | 4'-phosphopantetheinyl transferase PptT, COENZYME A, ISOPROPYL ALCOHOL, ... | | Authors: | Gavalda, S, Mourey, L, Pedelacq, J.D. | | Deposit date: | 2023-10-27 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Catalytic Cycle of Type II 4'-Phosphopantetheinyl Transferases
Acs Catalysis, 14, 2024
|
|
7BDW
 
 | | Crystal structure of mycobacterial PptAb in complex with ACP and compound 8918 | | Descriptor: | COENZYME A, MANGANESE (II) ION, N-(2,6-diethylphenyl)-N'-(N-ethylcarbamimidoyl)urea, ... | | Authors: | Carivenc, C, Nguyen, M.C, Mourey, L, Pedelacq, J.D. | | Deposit date: | 2020-12-22 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Phosphopantetheinyl transferase binding and inhibition by amidino-urea and hydroxypyrimidinethione compounds.
Sci Rep, 11, 2021
|
|
7BCZ
 
 | | Structure of the 4'-phosphopantetheinyl transferase PcpS from Pseudomonas aeruginosa | | Descriptor: | 4'-phosphopantetheinyl transferase EntD, COENZYME A, MAGNESIUM ION, ... | | Authors: | Carivenc, C, Mourey, L, Pedelacq, J.D. | | Deposit date: | 2020-12-21 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Phosphopantetheinyl transferase binding and inhibition by amidino-urea and hydroxypyrimidinethione compounds.
Sci Rep, 11, 2021
|
|
6HXU
 
 | | Crystal structure of Human RHOB Q63L in complex with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Rho-related GTP-binding protein RhoB | | Authors: | Soulie, S, Gence, R, Cabantous, S, Lajoie-Mazenc, I, Favre, G, Pedelacq, J.D. | | Deposit date: | 2018-10-18 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | A Targeted Protein Degradation Cell-Based Screening for Nanobodies Selective toward the Cellular RHOB GTP-Bound Conformation.
Cell Chem Biol, 26, 2019
|
|
5NJI
 
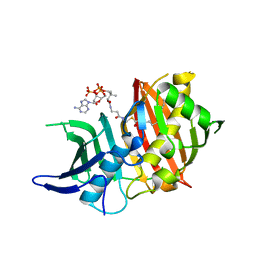 | | Structure of the dehydratase domain of PpsC from Mycobacterium tuberculosis in complex with C12:1-CoA | | Descriptor: | Phthiocerol/phenolphthiocerol synthesis polyketide synthase type I PpsC, ~{S}-[2-[3-[[(2~{R})-4-[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethyl] (~{E})-dodec-2-enethioate | | Authors: | Gavalda, S, Faille, A, Mourey, L, Pedelacq, J.D. | | Deposit date: | 2017-03-28 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights into Substrate Modification by Dehydratases from Type I Polyketide Synthases.
J. Mol. Biol., 429, 2017
|
|
5L84
 
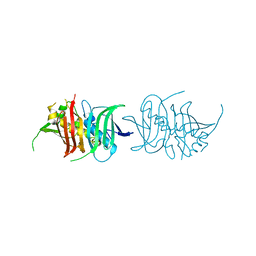 | |
5I0K
 
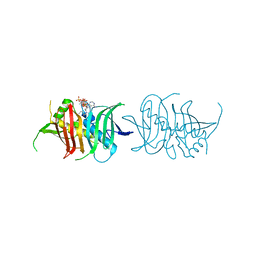 | |
1UHV
 
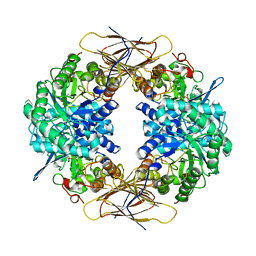 | | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase | | Descriptor: | 1,5-anhydro-2-deoxy-2-fluoro-D-xylitol, Beta-xylosidase | | Authors: | Yang, J.K, Yoon, H.J, Ahn, H.J, Il Lee, B, Pedelacq, J.D, Liong, E.C, Berendzen, J, Laivenieks, M, Vieille, C, Zeikus, G.J, Vocadlo, D.J, Withers, S.G, Suh, S.W. | | Deposit date: | 2003-07-11 | | Release date: | 2003-12-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase.
J.Mol.Biol., 335, 2004
|
|
1XVQ
 
 | | Crystal structure of thiol peroxidase from Mycobacterium tuberculosis | | Descriptor: | AMMONIUM ION, YTTRIUM (III) ION, thiol peroxidase | | Authors: | Rho, B.S, Pedelacq, J.D, Hung, L.W, Holton, J.M, Vigil, D, Kim, S.I, Park, M.S, Terwilliger, T.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-10-28 | | Release date: | 2004-12-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Functional and Structural Characterization of a Thiol Peroxidase from Mycobacterium tuberculosis.
J.Mol.Biol., 361, 2006
|
|
6QXR
 
 | | 4'-phosphopantetheinyl transferase PptAb from Mycobacterium abscessus at pH 8.5 with Mn2+ and CoA. | | Descriptor: | CALCIUM ION, COENZYME A, MANGANESE (II) ION, ... | | Authors: | Nguyen, M.C, Mourey, L, Pedelacq, J.D. | | Deposit date: | 2019-03-07 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Conformational flexibility of coenzyme A and its impact on the post-translational modification of acyl carrier proteins by 4'-phosphopantetheinyl transferases.
Febs J., 287, 2020
|
|
6QWU
 
 | | 4'-phosphopantetheinyl transferase PptAb from Mycobacterium abscessus at pH 5.5 with Mn2+ and CoA. | | Descriptor: | COENZYME A, DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, ... | | Authors: | Nguyen, M.C, Mourey, L, Pedelacq, J.D. | | Deposit date: | 2019-03-06 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Conformational flexibility of coenzyme A and its impact on the post-translational modification of acyl carrier proteins by 4'-phosphopantetheinyl transferases.
Febs J., 287, 2020
|
|
6QXQ
 
 | |
6RCX
 
 | | Mycobacterial 4'-phosphopantetheinyl transferase PptAb in complex with the ACP domain of PpsC. | | Descriptor: | CACODYLATE ION, COENZYME A, MANGANESE (II) ION, ... | | Authors: | Nguyen, M.C, Mourey, L, Pedelacq, J.D. | | Deposit date: | 2019-04-12 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational flexibility of coenzyme A and its impact on the post-translational modification of acyl carrier proteins by 4'-phosphopantetheinyl transferases.
Febs J., 287, 2020
|
|
6SGE
 
 | | Crystal structure of Human RHOB-GTP in complex with nanobody B6 | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Nanobody B6, ... | | Authors: | Soulie, S, Gence, R, Cabantous, S, Lajoie-Mazenc, I, Favre, G, Pedelacq, J.D. | | Deposit date: | 2019-08-04 | | Release date: | 2019-09-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Targeted Protein Degradation Cell-Based Screening for Nanobodies Selective toward the Cellular RHOB GTP-Bound Conformation.
Cell Chem Biol, 26, 2019
|
|
6QYG
 
 | |
6QYF
 
 | |
1AVB
 
 | | ARCELIN-1 FROM PHASEOLUS VULGARIS L | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ARCELIN-1, ... | | Authors: | Mourey, L, Pedelacq, J.D, Fabre, C, Rouge, P, Samama, J.P. | | Deposit date: | 1997-09-15 | | Release date: | 1998-10-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the arcelin-1 dimer from Phaseolus vulgaris at 1.9-A resolution.
J.Biol.Chem., 273, 1998
|
|
1BT5
 
 | | CRYSTAL STRUCTURE OF THE IMIPENEM INHIBITED TEM-1 BETA-LACTAMASE FROM ESCHERICHIA COLI | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, PROTEIN (BETA-LACTAMASE), SULFATE ION | | Authors: | Maveyraud, L, Mourey, L, Pedelacq, J.D, Guillet, V, Kotra, L.K, Mobashery, S, Samama, J.P. | | Deposit date: | 1998-09-02 | | Release date: | 1999-09-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Clinical Longevity of Carbapenem Antibiotics in the Face of Challenge by the Common Class A Beta-Lactamases from Antibiotic-Resistant Bacteria
J.Am.Chem.Soc., 120, 1998
|
|
