1Y9O
 
 | | 1H NMR Structure of Acylphosphatase from the hyperthermophile Sulfolobus Solfataricus | | Descriptor: | Acylphosphatase | | Authors: | Corazza, A, Rosano, C, Pagano, K, Alverdi, V, Esposito, G, Capanni, C, Bemporad, F, Plakoutsi, G, Stefani, M, Chiti, F, Zuccotti, S, Bolognesi, M, Viglino, P. | | Deposit date: | 2004-12-16 | | Release date: | 2005-11-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure, conformational stability, and enzymatic properties of acylphosphatase from the hyperthermophile Sulfolobus solfataricus
Proteins, 62, 2006
|
|
8SV0
 
 | |
8U36
 
 | |
1G90
 
 | |
7WH4
 
 | |
4FPP
 
 | | Bacterial phosphotransferase | | Descriptor: | GLYCEROL, MAGNESIUM ION, NICKEL (II) ION, ... | | Authors: | Fioravanti, A, Clantin, B, Dewitte, F, Lens, Z, Verger, A, Biondi, E, Villeret, V. | | Deposit date: | 2012-06-22 | | Release date: | 2012-09-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into ChpT, an essential dimeric histidine phosphotransferase regulating the cell cycle in Caulobacter crescentus.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
9C0Y
 
 | | Clathrin terminal domain complexed with Pitstop 2c | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Clathrin heavy chain 1, ... | | Authors: | Bulut, H, Horatscheck, A, Krauss, M, Santos, K.F, McCluskey, A, Wahl, C.W, Nazare, M, Haucke, V. | | Deposit date: | 2024-05-28 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Acute inhibition of clathrin-mediated endocytosis by next-generation small molecule inhibitors of clathrin function
To Be Published
|
|
9C0Z
 
 | | Clathrin terminal domain complexed with pitstop 2d | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Bulut, H, Horatscheck, A, Krauss, M, Santos, K.F, McCluskey, A, Wahl, C.W, Nazare, M, Haucke, V. | | Deposit date: | 2024-05-28 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Acute inhibition of clathrin-mediated endocytosis by next-generation small molecule inhibitors of clathrin function
To Be Published
|
|
6L6O
 
 | | Crystal structure of stabilized Rab5a GTPase domain from Leishmania donovani | | Descriptor: | ACETATE ION, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Arora, A, Zohib, M, Biswal, B.K, Maheshwari, D, Pal, R.K. | | Deposit date: | 2019-10-29 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the GDP-bound GTPase domain of Rab5a from Leishmania donovani.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6QX4
 
 | |
6HHU
 
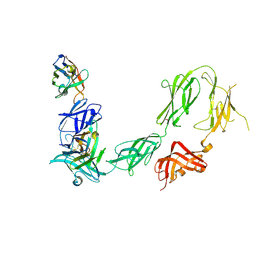 | |
4RP7
 
 | |
4RP6
 
 | |
4QXX
 
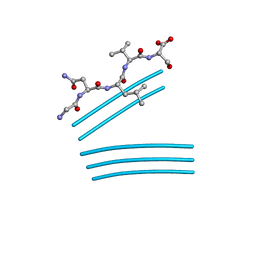 | |
2JTE
 
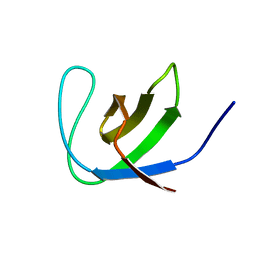 | | Third SH3 domain of CD2AP | | Descriptor: | CD2-associated protein | | Authors: | van Nuland, N.A.J, Ortega, R.J, Romero Romero, M, Ab, E, Ora, A, Lopez, M.O, Azuaga, A.I. | | Deposit date: | 2007-07-28 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The high resolution NMR structure of the third SH3 domain of CD2AP.
J.Biomol.Nmr, 39, 2007
|
|
5MJV
 
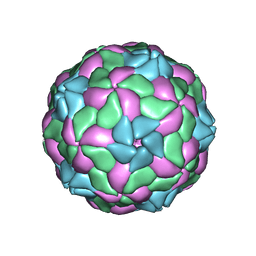 | | Rebuild and re-refined model for Human Parechovirus 1 | | Descriptor: | CAPSID SUBUNIT VP0, CAPSID SUBUNIT VP1, CAPSID SUBUNIT VP3, ... | | Authors: | Shakeel, S, Dykeman, E.C, White, S.J, Ora, A, Cockburn, J.J.B, Butcher, S.J, Stockley, P.G, Twarock, R. | | Deposit date: | 2016-12-01 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Genomic RNA folding mediates assembly of human parechovirus.
Nat Commun, 8, 2017
|
|
4UDF
 
 | | STRUCTURAL BASIS OF HUMAN PARECHOVIRUS NEUTRALIZATION BY HUMAN MONOCLONAL ANTIBODIES | | Descriptor: | HUMAN MONOCLONAL ANTIBODY, Protein VP0, Protein VP3 | | Authors: | Shakeel, S, Westerhuis, B.M, Ora, A, Koen, G, Bakker, A.Q, Claassen, Y, Beaumont, T, Wolthers, K.C, Butcher, S.J. | | Deposit date: | 2014-12-10 | | Release date: | 2015-07-22 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Structural Basis of Human Parechovirus Neutralization by Human Monoclonal Antibodies.
J. Virol., 89, 2015
|
|
3KK0
 
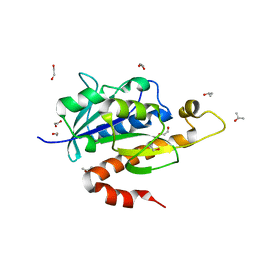 | | Crystal structure of partially folded intermediate state of peptidyl-tRNA hydrolase from Mycobacterium smegmatis | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kumar, A, Singh, N, Yadav, R, Sharma, S, Arora, A, Singh, T.P. | | Deposit date: | 2009-11-04 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structures of Fully-Folded Native and Partially-Folded Intermediate States of Peptidyl-tRNA Hydrolase from Mycobacterium smegmatis
To be Published
|
|
3KJZ
 
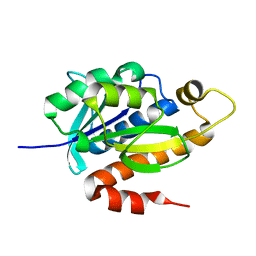 | | Crystal structure of native peptidyl-tRNA hydrolase from Mycobacterium smegmatis | | Descriptor: | Peptidyl-tRNA hydrolase | | Authors: | Kumar, A, Singh, N, Yadav, R, Prem Kumar, R, Sharma, S, Arora, A, Singh, T.P. | | Deposit date: | 2009-11-04 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of peptidyl-tRNA hydrolase from mycobacterium smegmatis reveals novel features related to enzyme dynamics.
Int J Biochem Mol Biol, 3, 2012
|
|
2KVK
 
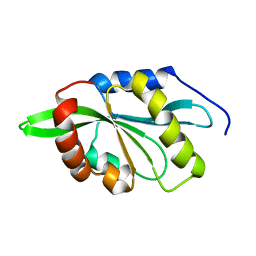 | | Solution structure of ADF/cofilin (LDCOF) from Leishmania donovani | | Descriptor: | Actin severing and dynamics regulatory protein | | Authors: | Pathak, P.P, Pulavarti, S.V, Jain, A, Sahasrabuddhe, A.A, Gupta, C.M, Arora, A. | | Deposit date: | 2010-03-16 | | Release date: | 2010-07-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of ADF/cofilin from Leishmania donovani
J.Struct.Biol., 172, 2010
|
|
2LBU
 
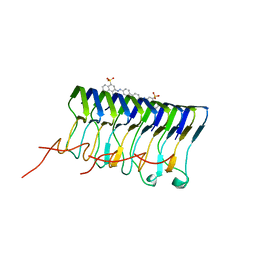 | | HADDOCK calculated model of Congo red bound to the HET-s amyloid | | Descriptor: | Small s protein, sodium 3,3'-(1E,1'E)-biphenyl-4,4'-diylbis(diazene-2,1-diyl)bis(4-aminonaphthalene-1-sulfonate) | | Authors: | Schutz, A.K, Soragni, A, Hornemann, S, Aguzzi, A, Ernst, M, Bockmann, A, Meier, B.H. | | Deposit date: | 2011-04-07 | | Release date: | 2011-06-01 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | The Amyloid-Congo Red Interface at Atomic Resolution.
Angew.Chem.Int.Ed.Engl., 2011
|
|
2KGY
 
 | | Solution structure of Rv0603 protein from Mycobacterium tuberculosis H37Rv | | Descriptor: | POSSIBLE EXPORTED PROTEIN | | Authors: | Tripathi, S, Pulavarti, S.V.S.R.K, Pathak, P.P, Meher, A.K, Jain, A, Arora, A. | | Deposit date: | 2009-03-23 | | Release date: | 2010-03-31 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Rv0603 protein from Mycobacterium tuberculosis H37Rv
To be Published
|
|
2L72
 
 | | Solution structure and dynamics of ADF from Toxoplasma gondii (TgADF) | | Descriptor: | Actin depolymerizing factor, putative | | Authors: | Pathak, P.P, Shukla, V.K, Yadav, R, Jain, A, Srivastava, S, Tripathi, S, Pulavarti, S.V.S.R.K, Arora, A. | | Deposit date: | 2010-12-01 | | Release date: | 2011-08-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of ADF from Toxoplasma gondii
J.Struct.Biol., 176, 2011
|
|
9C20
 
 | | The Sialidase NanJ in complex with Neu5,9Ac | | Descriptor: | 1,2-ETHANEDIOL, 9-O-acetyl-5-acetamido-3,5-dideoxy-D-glycero-alpha-D-galacto-non-2-ulopyranosonic acid, exo-alpha-sialidase | | Authors: | Medley, B.J, Low, K.E, Garber, J.M, Gray, T.E, Leeann, L.L, Fordwour, O.B, Inglis, G.D, Boons, G.J, Zandberg, W.F, Abbott, W, Boraston, A. | | Deposit date: | 2024-05-30 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.699 Å) | | Cite: | NanJ sialidase in complex with Neu5,9Ac
To Be Published
|
|
2AEB
 
 | | Crystal structure of human arginase I at 1.29 A resolution and exploration of inhibition in immune response. | | Descriptor: | 2(S)-AMINO-6-BORONOHEXANOIC ACID, Arginase 1, MANGANESE (II) ION | | Authors: | Di Costanzo, L, Sabio, G, Mora, A, Rodriguez, P.C, Ochoa, A.C, Centeno, F, Christianson, D.W. | | Deposit date: | 2005-07-21 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Crystal structure of human arginase I at 1.29 A resolution and exploration of inhibition in the immune response.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
