1VF9
 
 | | Solution Structure Of Human Trf2 | | Descriptor: | Telomeric repeat binding factor 2 | | Authors: | Nishimura, Y, Hanaoka, S. | | Deposit date: | 2004-04-12 | | Release date: | 2005-05-17 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Comparison between TRF2 and TRF1 of their telomeric DNA-bound structures and DNA-binding activities
Protein Sci., 14, 2005
|
|
1VFC
 
 | | Solution Structure Of The DNA Complex Of Human Trf2 | | Descriptor: | Short C-rich starnd, Short G-rich strand, Telomeric repeat binding factor 2 | | Authors: | Nishimura, Y, Hanaoka, S. | | Deposit date: | 2004-04-12 | | Release date: | 2005-05-17 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Comparison between TRF2 and TRF1 of their telomeric DNA-bound structures and DNA-binding activities
Protein Sci., 14, 2005
|
|
3D79
 
 | | Crystal structure of hypothetical protein PH0734.1 from hyperthermophilic archaea Pyrococcus horikoshii OT3 | | Descriptor: | Putative uncharacterized protein PH0734 | | Authors: | Nishimura, Y, Miyazono, K, Sawano, Y, Makino, T, Nagata, K, Tanokura, M. | | Deposit date: | 2008-05-20 | | Release date: | 2008-12-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of hypothetical protein PH0734.1 from hyperthermophilic archaea Pyrococcus horikoshii OT3.
Proteins, 73, 2008
|
|
8I53
 
 | |
1MBH
 
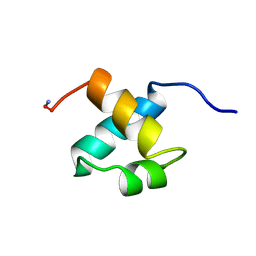 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 2 | | Descriptor: | C-MYB | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-09-15 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
1MBE
 
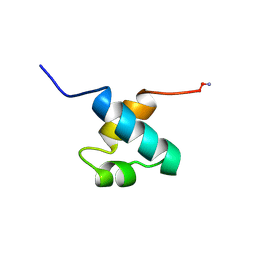 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 1 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-07-31 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
1MBG
 
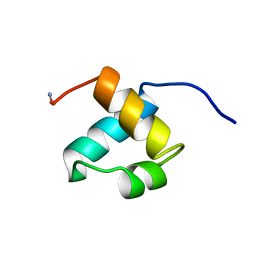 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 2 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-07-31 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
1MBJ
 
 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 3 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-07-31 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
1MBK
 
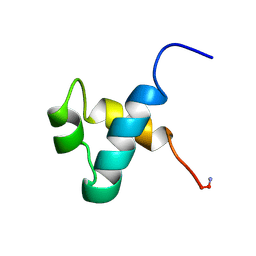 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 3 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-07-31 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
1MBF
 
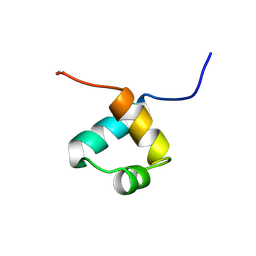 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 1 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-07-31 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
3J3Z
 
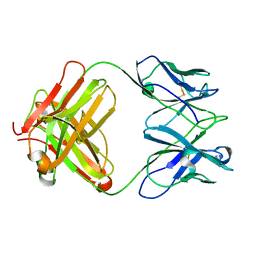 | | Structure of MA28-7 neutralizing antibody Fab fragment from electron cryo-microscopy of enterovirus 71 complexed with a Fab fragment | | Descriptor: | MA28-7 neutralizing antibody heavy chain, MA28-7 neutralizing antibody light chain | | Authors: | Lee, H, Cifuente, J.O, Ashley, R.E, Conway, J.F, Makhov, A.M, Tano, Y, Shimizu, H, Nishimura, Y, Hafenstein, S. | | Deposit date: | 2013-05-21 | | Release date: | 2013-08-28 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (23.4 Å) | | Cite: | A strain-specific epitope of enterovirus 71 identified by cryo-electron microscopy of the complex with fab from neutralizing antibody.
J.Virol., 87, 2013
|
|
1MSE
 
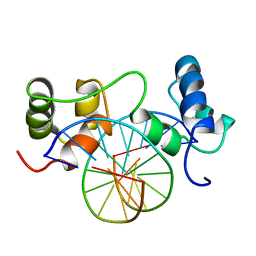 | | SOLUTION STRUCTURE OF A SPECIFIC DNA COMPLEX OF THE MYB DNA-BINDING DOMAIN WITH COOPERATIVE RECOGNITION HELICES | | Descriptor: | C-Myb DNA-Binding Domain, DNA (5'-D(*AP*TP*GP*TP*GP*TP*GP*TP*CP*AP*GP*TP*TP*AP*GP*G)-3'), DNA (5'-D(*CP*CP*TP*AP*AP*CP*TP*GP*AP*CP*AP*CP*AP*CP*AP*T)-3') | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Sekikawa, A, Inoue, T, Kanai, H, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-01-24 | | Release date: | 1995-03-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices.
Cell(Cambridge,Mass.), 79, 1994
|
|
1MSF
 
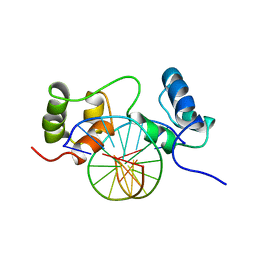 | | SOLUTION STRUCTURE OF A SPECIFIC DNA COMPLEX OF THE MYB DNA-BINDING DOMAIN WITH COOPERATIVE RECOGNITION HELICES | | Descriptor: | C-Myb DNA-Binding Domain, DNA (5'-D(*AP*TP*GP*TP*GP*TP*GP*TP*CP*AP*GP*TP*TP*AP*GP*G)-3'), DNA (5'-D(*CP*CP*TP*AP*AP*CP*TP*GP*AP*CP*AP*CP*AP*CP*AP*T)-3') | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Sekikawa, A, Inoue, T, Kanai, H, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-01-24 | | Release date: | 1995-03-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices.
Cell(Cambridge,Mass.), 79, 1994
|
|
5Y95
 
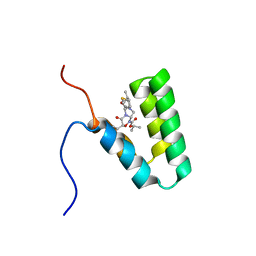 | | Haddock model of mSIN3B PAH1 domain | | Descriptor: | Paired amphipathic helix protein Sin3b, propan-2-yl (3R,6S,9aS)-3-ethyl-8-(3-methylbutyl)-6-(2-methylsulfanylethyl)-4,7-bis(oxidanylidene)-9,9a-dihydro-6H-pyrazino[2,1-c][1,2,4]oxadiazine-1-carboxylate | | Authors: | Kurita, J, Hirao, Y, Nishimura, Y. | | Deposit date: | 2017-08-22 | | Release date: | 2017-10-04 | | Last modified: | 2025-03-12 | | Method: | SOLUTION NMR | | Cite: | A mimetic of the mSin3-binding helix of NRSF/REST ameliorates abnormal pain behavior in chronic pain models.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
2CZY
 
 | | Solution structure of the NRSF/REST-mSin3B PAH1 complex | | Descriptor: | Paired amphipathic helix protein Sin3b, transcription factor REST (version 3) | | Authors: | Nomura, M, Uda-Tochio, H, Murai, K, Mori, N, Nishimura, Y. | | Deposit date: | 2005-07-20 | | Release date: | 2005-12-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Neural Repressor NRSF/REST Binds the PAH1 Domain of the Sin3 Corepressor by Using its Distinct Short Hydrophobic Helix
J.Mol.Biol., 354, 2005
|
|
1BA5
 
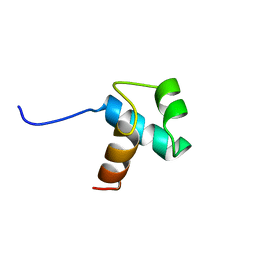 | | DNA-BINDING DOMAIN OF HUMAN TELOMERIC PROTEIN, HTRF1, NMR, 18 STRUCTURES | | Descriptor: | HTRF1 | | Authors: | Nishikawa, T, Nagadoi, A, Yoshimura, S, Aimoto, S, Nishimura, Y. | | Deposit date: | 1998-04-22 | | Release date: | 1999-04-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA-binding domain of human telomeric protein, hTRF1.
Structure, 6, 1998
|
|
1BHI
 
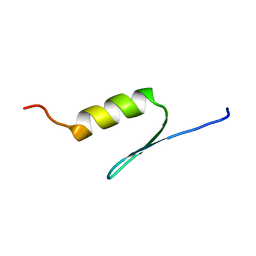 | | STRUCTURE OF TRANSACTIVATION DOMAIN OF CRE-BP1/ATF-2, NMR, 20 STRUCTURES | | Descriptor: | CRE-BP1 | | Authors: | Nagadoi, A, Nakazawa, K, Uda, H, Maekawa, T, Ishii, S, Nishimura, Y. | | Deposit date: | 1998-06-09 | | Release date: | 1999-06-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the transactivation domain of ATF-2 comprising a zinc finger-like subdomain and a flexible subdomain.
J.Mol.Biol., 287, 1999
|
|
1D8J
 
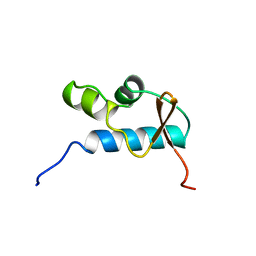 | | SOLUTION STRUCTURE OF THE CENTRAL CORE DOMAIN OF TFIIE BETA | | Descriptor: | GENERAL TRANSCRIPTION FACTOR TFIIE-BETA | | Authors: | Okuda, M, Watanabe, Y, Okamura, H, Hanaoka, F, Ohkuma, Y, Nishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-10-25 | | Release date: | 2000-04-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the central core domain of TFIIEbeta with a novel double-stranded DNA-binding surface.
EMBO J., 19, 2000
|
|
1D8K
 
 | | SOLUTION STRUCTURE OF THE CENTRAL CORE DOMAIN OF TFIIE BETA | | Descriptor: | GENERAL TRANSCRIPTION FACTOR TFIIE-BETA | | Authors: | Okuda, M, Watanabe, Y, Okamura, H, Hanaoka, F, Ohkuma, Y, Nishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-10-25 | | Release date: | 2000-04-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the central core domain of TFIIEbeta with a novel double-stranded DNA-binding surface.
EMBO J., 19, 2000
|
|
1PRU
 
 | | PURINE REPRESSOR DNA-BINDING DOMAIN DNA BINDING | | Descriptor: | PURINE REPRESSOR | | Authors: | Nagadoi, A, Morikawa, S, Nakamura, H, Enari, M, Kobayashi, K, Yamamoto, H, Sampei, G, Mizobuchi, K, Schumacher, M.A, Brennan, R.G, Nishimura, Y. | | Deposit date: | 1995-05-08 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural comparison of the free and DNA-bound forms of the purine repressor DNA-binding domain.
Structure, 3, 1995
|
|
1PRV
 
 | | PURINE REPRESSOR DNA-BINDING DOMAIN DNA BINDING | | Descriptor: | PURINE REPRESSOR | | Authors: | Nagadoi, A, Morikawa, S, Nakamura, H, Enari, M, Kobayashi, K, Yamamoto, H, Sampei, G, Mizobuchi, K, Schumacher, M.A, Brennan, R.G, Nishimura, Y. | | Deposit date: | 1995-05-08 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural comparison of the free and DNA-bound forms of the purine repressor DNA-binding domain.
Structure, 3, 1995
|
|
1QQI
 
 | | SOLUTION STRUCTURE OF THE DNA-BINDING AND TRANSACTIVATION DOMAIN OF PHOB FROM ESCHERICHIA COLI | | Descriptor: | PHOSPHATE REGULON TRANSCRIPTIONAL REGULATORY PROTEIN PHOB | | Authors: | Okamura, H, Hanaoka, S, Nagadoi, A, Makino, K, Nishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-06-07 | | Release date: | 2000-06-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural comparison of the PhoB and OmpR DNA-binding/transactivation domains and the arrangement of PhoB molecules on the phosphate box.
J.Mol.Biol., 295, 2000
|
|
7DTI
 
 | |
2DY7
 
 | |
1VD4
 
 | | Solution structure of the zinc finger domain of TFIIE alpha | | Descriptor: | Transcription initiation factor IIE, alpha subunit, ZINC ION | | Authors: | Okuda, M, Tanaka, A, Arai, Y, Satoh, M, Okamura, H, Nagadoi, A, Hanaoka, F, Ohkuma, Y, Nishimura, Y. | | Deposit date: | 2004-03-18 | | Release date: | 2004-10-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A novel zinc finger structure in the large subunit of human general transcription factor TFIIE.
J.Biol.Chem., 279, 2004
|
|
