4OVO
 
 | |
3OVO
 
 | |
1HUC
 
 | |
5BNJ
 
 | | CDK8/CYCC IN COMPLEX WITH 8-{3-Chloro-5-[4-(1-methyl-1H-pyrazol-4-yl)-phenyl]-pyridin- 4-yl}-2,8-diaza-spiro[4.5]decan-1-one | | Descriptor: | 8-{3-chloro-5-[4-(1-methyl-1H-pyrazol-4-yl)phenyl]pyridin-4-yl}-2,8-diazaspiro[4.5]decan-1-one, Cyclin-C, Cyclin-dependent kinase 8, ... | | Authors: | Musil, D, Blagg, J, Wienke, D. | | Deposit date: | 2015-05-26 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | A selective chemical probe for exploring the role of CDK8 and CDK19 in human disease.
Nat.Chem.Biol., 11, 2015
|
|
7OCV
 
 | | Human TNKS1 in complex with 3-[4-(1-Hydroxy-1-methyl-ethyl)-phenyl]-6-methyl-2H-pyrrolo[1,2-a]pyrazin-1-one | | Descriptor: | 6-methyl-3-[4-(2-oxidanylpropan-2-yl)phenyl]-4~{H}-pyrrolo[1,2-a]pyrazin-1-one, ACETATE ION, Poly [ADP-ribose] polymerase, ... | | Authors: | Musil, D, Lehmann, M, Buchstaller, H.-P. | | Deposit date: | 2021-04-28 | | Release date: | 2021-07-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.432 Å) | | Cite: | Optimization of a Screening Hit toward M2912, an Oral Tankyrase Inhibitor with Antitumor Activity in Colorectal Cancer Models.
J.Med.Chem., 64, 2021
|
|
6TXZ
 
 | | FAB PART OF M6903 IN COMPLEX WITH HUMAN TIM3 | | Descriptor: | Fab H, Fab L, Hepatitis A virus cellular receptor 2 | | Authors: | Musil, D, Sood, V. | | Deposit date: | 2020-01-15 | | Release date: | 2020-04-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Identification and characterization of M6903, an antagonistic anti-TIM-3 monoclonal antibody.
Oncoimmunology, 9, 2020
|
|
9SDI
 
 | |
7ZJQ
 
 | |
9FZA
 
 | |
9GAX
 
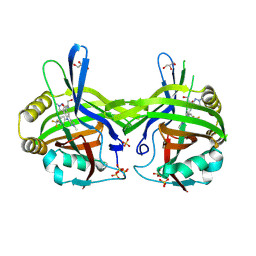 | | TEAD1 in complex with a reversible inhibitor N-[(1S)-2-hydroxy-1-(1-methyl-1H-pyrazol-3-yl)ethyl]-2-methyl-8-[4-(trifluoromethyl)phenyl]-2H,8H-pyrazolo[3,4-b]indole-5-carboxamide | | Descriptor: | 2-methyl-~{N}-[(1~{S})-1-(1-methylpyrazol-3-yl)-2-oxidanyl-ethyl]-4-[4-(trifluoromethyl)phenyl]pyrazolo[3,4-b]indole-7-carboxamide, GLYCEROL, SULFATE ION, ... | | Authors: | Musil, D, Freire, F. | | Deposit date: | 2024-07-29 | | Release date: | 2025-01-22 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | MoA Studies of the TEAD P-Site Binding Ligand MSC-4106 and Its Optimization to TEAD1-Selective Amide M3686.
J.Med.Chem., 68, 2025
|
|
5FGK
 
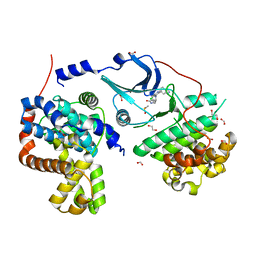 | | CDK8-CYCC IN COMPLEX WITH 8-[3-(3-Amino-1H-indazol-6-yl)-5-chloro- pyridine-4-yl]-2,8-diaza-spiro[4.5]decan-1-one | | Descriptor: | 1,2-ETHANEDIOL, 8-[3-(3-azanyl-2~{H}-indazol-6-yl)-5-chloranyl-pyridin-4-yl]-2,8-diazaspiro[4.5]decan-1-one, Cyclin-C, ... | | Authors: | Musil, D, Blagg, J, Mallinger, A. | | Deposit date: | 2015-12-20 | | Release date: | 2016-02-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Discovery of Potent, Selective, and Orally Bioavailable Small-Molecule Modulators of the Mediator Complex-Associated Kinases CDK8 and CDK19.
J.Med.Chem., 59, 2016
|
|
6HHR
 
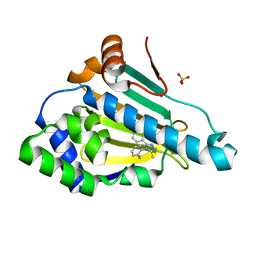 | | Hsp90 in complex with 5-(2,4-Dihydroxy-phenyl)-4-(2-fluoro-phenyl)-2,4-dihydro-[1,2,4]triazole-3-thione | | Descriptor: | 3-[2,4-bis(oxidanyl)phenyl]-4-(2-fluorophenyl)-1~{H}-1,2,4-triazole-5-thione, Heat shock protein HSP 90-alpha, SULFATE ION | | Authors: | Musil, D, Lehman, M, Eggenweiler, H.-M. | | Deposit date: | 2018-08-29 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Predicting Residence Time and Drug Unbinding Pathway through Scaled Molecular Dynamics.
J.Chem.Inf.Model., 59, 2019
|
|
7B12
 
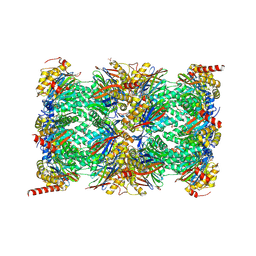 | | HUMAN IMMUNOPROTEASOME 20S PARTICLE IN COMPLEX WITH [2-(3-ethylphenyl)-1-[(2S)-3-phenyl-2-[(pyrazin-2-yl)formamido]propanamido]ethyl]boronic acid | | Descriptor: | ((R)-2-(3-ethylphenyl)-1-((S)-3-phenyl-2-(pyrazine-2-carboxamido)propanamido)ethyl)boronic acid, Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, ... | | Authors: | Musil, D, Klein, M, Crosignani, S. | | Deposit date: | 2020-11-23 | | Release date: | 2021-12-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structure-Based Optimization and Discovery of M3258, a Specific Inhibitor of the Immunoproteasome Subunit LMP7 ( beta 5i).
J.Med.Chem., 64, 2021
|
|
9S64
 
 | | Human TEAD1 in complex with 2-(4-chloro-3-{3-methyl-5-[4-(trifluoromethyl)phenoxy]phenyl}-1H-pyrrolo[3,2-c]pyridin-1-yl)ethan-1-ol | | Descriptor: | 2-[4-chloranyl-3-[3-methyl-5-[4-(trifluoromethyl)phenoxy]phenyl]pyrrolo[3,2-c]pyridin-1-yl]ethanol, CHLORIDE ION, GLYCEROL, ... | | Authors: | Musil, D, Freire, F. | | Deposit date: | 2025-07-30 | | Release date: | 2026-02-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Phenotypic Hit Identification and Optimization of Novel Pan-TEAD and Subtype-Selective Inhibitors.
J.Med.Chem., 68, 2025
|
|
9FXF
 
 | |
9FWW
 
 | | Human NKp30 in complex with a VHH variant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Natural cytotoxicity triggering receptor 3, VHH, ... | | Authors: | Musil, D, Freire, F. | | Deposit date: | 2024-07-01 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.844 Å) | | Cite: | On the humanization of VHHs: Prospective case studies, experimental and computational characterization of structural determinants for functionality.
Protein Sci., 33, 2024
|
|
8CK3
 
 | | STRUCTURE OF HIF2A-ARNT HETERODIMER IN COMPLEX WITH (S)-1-(3,5-Difluoro-phenyl)-5,5-difluoro-3-methanesulfonyl-5,6-dihydro-4H-cyclopenta[c]thiophen-4-ol | | Descriptor: | (4~{S})-1-[3,5-bis(fluoranyl)phenyl]-5,5-bis(fluoranyl)-3-methylsulfonyl-4,6-dihydrocyclopenta[c]thiophen-4-ol, Aryl hydrocarbon receptor nuclear translocator, DIMETHYL SULFOXIDE, ... | | Authors: | Musil, D, Lehmannn, M, Diehl, L. | | Deposit date: | 2023-02-14 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.707 Å) | | Cite: | Discovery of Cycloalkyl[ c ]thiophenes as Novel Scaffolds for Hypoxia-Inducible Factor-2 alpha Inhibitors.
J.Med.Chem., 66, 2023
|
|
8CK8
 
 | |
8CK4
 
 | | STRUCTURE OF HIF2A-ARNT HETERODIMER IN COMPLEX WITH (4S)-1-(3,5-difluorophenyl)-5,5-difluoro-3-methanesulfonyl-4,5,6,7-tetrahydro-2-benzothiophen-4-ol | | Descriptor: | (4~{S})-1-[3,5-bis(fluoranyl)phenyl]-5,5-bis(fluoranyl)-3-methylsulfonyl-6,7-dihydro-4~{H}-2-benzothiophen-4-ol, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Musil, D. | | Deposit date: | 2023-02-14 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Discovery of Cycloalkyl[ c ]thiophenes as Novel Scaffolds for Hypoxia-Inducible Factor-2 alpha Inhibitors.
J.Med.Chem., 66, 2023
|
|
6QXU
 
 | | Human TNKS1 in complex with 6,8-Difluoro-2-[4-(1-hydroxy-1-methyl-ethyl)-phenyl]-3H-quinazolin-4-one | | Descriptor: | 6,8-bis(fluoranyl)-2-[4-(2-oxidanylpropan-2-yl)phenyl]-3~{H}-quinazolin-4-one, BETA-MERCAPTOETHANOL, Poly [ADP-ribose] polymerase tankyrase-1, ... | | Authors: | Musil, D, Lehmann, D, Buchstaller, H.-P. | | Deposit date: | 2019-03-08 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Discovery and Optimization of 2-Arylquinazolin-4-ones into a Potent and Selective Tankyrase Inhibitor Modulating Wnt Pathway Activity.
J.Med.Chem., 62, 2019
|
|
6ELP
 
 | |
6EL5
 
 | | Estimation of relative drug-target residence times by random acceleration molecular dynamics simulation | | Descriptor: | 8-(2-CHLORO-3,4,5-TRIMETHOXY-BENZYL)-2-FLUORO-9-PENT-4-YLNYL-9H-PURIN-6-YLAMINE, Heat shock protein HSP 90-alpha | | Authors: | Musil, D, Lehmann, M, Eggenweiler, H.-M. | | Deposit date: | 2017-09-27 | | Release date: | 2018-05-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Estimation of Drug-Target Residence Times by tau-Random Acceleration Molecular Dynamics Simulations.
J Chem Theory Comput, 14, 2018
|
|
6EI5
 
 | |
6QEG
 
 | | CRYSTAL STRUCTURE OF HUMAN METHIONINE AMINOPEPTIDASE-2 IN COMPLEX WITH AN INHIBITOR 2-Oxo-1-phenyl-pyrrolidine-3-carboxylic acid (2-thiophen-2-yl-ethyl)-amide | | Descriptor: | (3~{S})-3-oxidanyl-2-oxidanylidene-1-phenyl-~{N}-(2-thiophen-2-ylethyl)pyrrolidine-3-carboxamide, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Musil, D, Heinrich, T, Lehmann, M. | | Deposit date: | 2019-01-07 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Discovery and Structure-Based Optimization of Next-Generation Reversible Methionine Aminopeptidase-2 (MetAP-2) Inhibitors.
J.Med.Chem., 62, 2019
|
|
6QEI
 
 | | CRYSTAL STRUCTURE OF HUMAN METHIONINE AMINOPEPTIDASE-2 IN COMPLEX WITH AN INHIBITOR 5,6-Difluoro-3-(2-isopropoxy-4-piperazin-1-yl-phenyl)-1H-indole-2-carboxylic acid amide | | Descriptor: | 1,2-ETHANEDIOL, 5,6-bis(fluoranyl)-3-(4-piperazin-1-yl-2-propan-2-yloxy-phenyl)-1~{H}-indole-2-carboxamide, DIMETHYL SULFOXIDE, ... | | Authors: | Musil, D, Heinrich, T, Lehmann, M. | | Deposit date: | 2019-01-07 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery and Structure-Based Optimization of Next-Generation Reversible Methionine Aminopeptidase-2 (MetAP-2) Inhibitors.
J.Med.Chem., 62, 2019
|
|
