1BC7
 
 | | SERUM RESPONSE FACTOR ACCESSORY PROTEIN 1A (SAP-1)/DNA COMPLEX | | Descriptor: | DNA (5'-D(*CP*AP*CP*AP*TP*CP*CP*TP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*AP*GP*GP*AP*TP*GP*TP*G)-3'), PROTEIN (ETS-DOMAIN PROTEIN) | | Authors: | Mo, Y, Vaessen, B, Johnston, K, Marmorstein, R. | | Deposit date: | 1998-05-05 | | Release date: | 1999-01-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structures of SAP-1 bound to DNA targets from the E74 and c-fos promoters: insights into DNA sequence discrimination by Ets proteins.
Mol.Cell, 2, 1998
|
|
1BC8
 
 | | STRUCTURES OF SAP-1 BOUND TO DNA SEQUENCES FROM THE E74 AND C-FOS PROMOTERS PROVIDE INSIGHTS INTO HOW ETS PROTEINS DISCRIMINATE BETWEEN RELATED DNA TARGETS | | Descriptor: | DNA (5'-D(*AP*AP*CP*TP*TP*CP*CP*GP*GP*T)-3'), DNA (5'-D(*TP*AP*CP*CP*GP*GP*AP*AP*GP*T)-3'), PROTEIN (SAP-1 ETS DOMAIN), ... | | Authors: | Mo, Y, Vaessen, B, Johnston, K, Marmorstein, R. | | Deposit date: | 1998-05-05 | | Release date: | 1998-12-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structures of SAP-1 bound to DNA targets from the E74 and c-fos promoters: insights into DNA sequence discrimination by Ets proteins.
Mol.Cell, 2, 1998
|
|
1DUX
 
 | | ELK-1/DNA STRUCTURE REVEALS HOW RESIDUES DISTAL FROM DNA-BINDING SURFACE AFFECT DNA-RECOGNITION | | Descriptor: | DNA (5'-D(*AP*CP*AP*CP*TP*TP*CP*CP*GP*GP*TP*CP*A)-3'), DNA (5'-D(*TP*GP*AP*CP*CP*GP*GP*AP*AP*GP*TP*GP*T)-3'), ETS-DOMAIN PROTEIN ELK-1 | | Authors: | Mo, Y, Vaessen, B, Johnston, K, Marmorstein, R. | | Deposit date: | 2000-01-19 | | Release date: | 2000-04-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the elk-1-DNA complex reveals how DNA-distal residues affect ETS domain recognition of DNA.
Nat.Struct.Biol., 7, 2000
|
|
1K6O
 
 | | Crystal Structure of a Ternary SAP-1/SRF/c-fos SRE DNA Complex | | Descriptor: | 5'-D(*CP*AP*CP*AP*GP*GP*AP*TP*GP*TP*CP*CP*AP*TP*AP*TP*TP*AP*GP*GP*AP*CP*A)-3', 5'-D(*TP*GP*TP*CP*CP*TP*AP*AP*TP*AP*TP*GP*GP*AP*CP*AP*TP*CP*CP*TP*GP*TP*G)-3', ETS-domain protein ELK-4, ... | | Authors: | Mo, Y, Ho, W, Johnston, K, Marmorstein, R. | | Deposit date: | 2001-10-16 | | Release date: | 2002-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Crystal structure of a ternary SAP-1/SRF/c-fos SRE DNA complex.
J.Mol.Biol., 314, 2001
|
|
6IM5
 
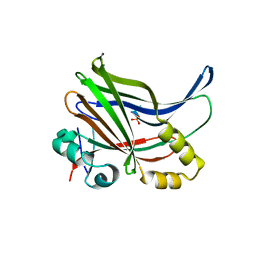 | | YAP-binding domain of human TEAD1 | | Descriptor: | PHOSPHATE ION, Transcriptional enhancer factor TEF-1 | | Authors: | Mo, Y, Lee, H.S, Kim, S.J, Ku, B. | | Deposit date: | 2018-10-22 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Crystal Structure of the YAP-binding Domain of Human TEAD1
Bull.Korean Chem.Soc., 40, 2019
|
|
3PS5
 
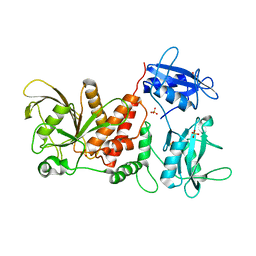 | | Crystal structure of the full-length Human Protein Tyrosine Phosphatase SHP-1 | | Descriptor: | SULFATE ION, Tyrosine-protein phosphatase non-receptor type 6 | | Authors: | Wang, W, Liu, L, Song, X, Mo, Y, Komma, C, Bellamy, H.D, Zhao, Z.J, Zhou, G.W. | | Deposit date: | 2010-11-30 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of human protein tyrosine phosphatase SHP-1 in the open conformation.
J.Cell.Biochem., 112, 2011
|
|
1QSR
 
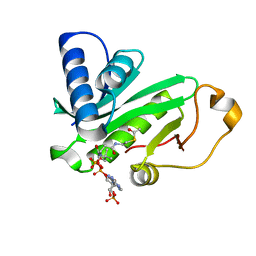 | | CRYSTAL STRUCTURE OF TETRAHYMENA GCN5 WITH BOUND ACETYL-COENZYME A | | Descriptor: | ACETYL COENZYME *A, TGCN5 HISTONE ACETYL TRANSFERASE | | Authors: | Rojas, J.R, Trievel, R.C, Zhou, J, Mo, Y, Li, X, Berger, S.L, David Allis, C, Marmorstein, R. | | Deposit date: | 1999-06-23 | | Release date: | 1999-09-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide.
Nature, 401, 1999
|
|
1QST
 
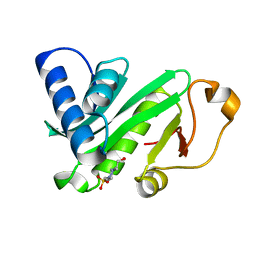 | | CRYSTAL STRUCTURE OF TETRAHYMENA GCN5 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, TGCN5 HISTONE ACETYL TRANSFERASE | | Authors: | Rojas, J.R, Trievel, R.C, Zhou, J, Mo, Y, Li, X, Berger, S.L, David Allis, C, Marmorstein, R. | | Deposit date: | 1999-06-23 | | Release date: | 1999-09-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide.
Nature, 401, 1999
|
|
1QSN
 
 | | CRYSTAL STRUCTURE OF TETRAHYMENA GCN5 WITH BOUND COENZYME A AND HISTONE H3 PEPTIDE | | Descriptor: | COENZYME A, HISTONE H3, TGCN5 HISTONE ACETYL TRANSFERASE | | Authors: | Rojas, J.R, Trievel, R.C, Zhou, J, Mo, Y, Li, X, Berger, S.L, David Allis, C, Marmorstein, R. | | Deposit date: | 1999-06-22 | | Release date: | 1999-09-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide.
Nature, 401, 1999
|
|
1DVJ
 
 | | CRYSTAL STRUCTURE OF OROTIDINE MONOPHOSPHATE DECARBOXYLASE COMPLEXED WITH 6-AZAUMP | | Descriptor: | 6-AZA URIDINE 5'-MONOPHOSPHATE, OROTIDINE 5'-PHOSPHATE DECARBOXYLASE | | Authors: | Wu, N, Mo, Y, Gao, J, Pai, E.F. | | Deposit date: | 2000-03-30 | | Release date: | 2000-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Electrostatic stress in catalysis: structure and mechanism of the enzyme orotidine monophosphate decarboxylase.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1DV7
 
 | |
7BTG
 
 | | Crystal structure of DARP, drosophila arginine phosphatase | | Descriptor: | CHLORIDE ION, GEO10716p1, PHOSPHATE ION | | Authors: | Lee, H.S, Mo, Y, Ku, B, Kim, S.J. | | Deposit date: | 2020-04-01 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.188 Å) | | Cite: | Structural and Biochemical Characterization of the Two Drosophila Low Molecular Weight-Protein Tyrosine Phosphatases DARP and Primo-1.
Mol.Cells, 43, 2020
|
|
7Y1Q
 
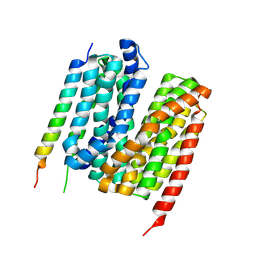 | | 5.0 angstrom cryo-EM structure of transmembrane regions of mouse Basigin/MCT1 in complex with antibody 6E7F1 | | Descriptor: | Isoform 2 of Basigin, Monocarboxylate transporter 1 | | Authors: | Zhang, H, Yang, X, Xue, Y, Huang, Y, Mo, X, Zhang, H, Li, N, Gao, N, Li, X, Wang, S, Gao, Y, Liao, J. | | Deposit date: | 2022-06-08 | | Release date: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (5.03 Å) | | Cite: | Allosteric modulation of monocarboxylate transporters 1 and 4 by targeting their chaperon Basigin-2
To Be Published
|
|
7Y1B
 
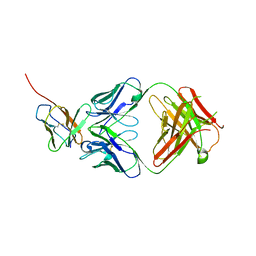 | | 3.2 angstrom cryo-EM structure of extracellular region of mouse Basigin-2 in complex with the Fab fragment of antibody 6E7F1 | | Descriptor: | Heavy chain of 6E7F1, Isoform 2 of Basigin, Light chain of 6E7F1 | | Authors: | Zhang, H, Yang, X, Xue, Y, Huang, Y, Mo, X, Zhang, H, Li, N, Gao, N, Li, X, Wang, S, Gao, Y, Liao, J. | | Deposit date: | 2022-06-08 | | Release date: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Allosteric modulation of monocarboxylate transporters 1 and 4 by targeting their chaperon Basigin
To Be Published
|
|
7CUY
 
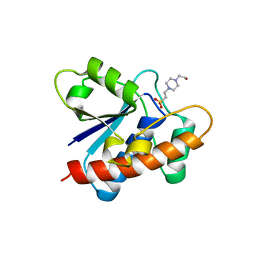 | | Crystal structure of Primo-1 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Low molecular weight phosphotyrosine protein phosphatase 1 | | Authors: | Lee, H.S, Kim, S.J, Ku, B. | | Deposit date: | 2020-08-25 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.081 Å) | | Cite: | Structural and Biochemical Characterization of the Two Drosophila Low Molecular Weight-Protein Tyrosine Phosphatases DARP and Primo-1.
Mol.Cells, 43, 2020
|
|
