7A6G
 
 | |
3N5F
 
 | | Crystal Structure of L-N-carbamoylase from Geobacillus stearothermophilus CECT43 | | Descriptor: | CACODYLATE ION, COBALT (II) ION, ISOPROPYL ALCOHOL, ... | | Authors: | Garcia-Pino, A, Martinez-Rodriguez, S, Gavira, J.A. | | Deposit date: | 2010-05-25 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Mutational and structural analysis of L-N-carbamoylase reveals new insights into a peptidase m20/m25/m40 family member.
J.Bacteriol., 194, 2012
|
|
8BV9
 
 | | Acylphosphatase from E. coli | | Descriptor: | 1,2-ETHANEDIOL, Acylphosphatase, GLYCEROL, ... | | Authors: | Gavira, J.A, Martinez-Rodriguez, S. | | Deposit date: | 2023-01-18 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | First 3-D structural evidence of a native-like intertwined dimer in the acylphosphatase family.
Biochem.Biophys.Res.Commun., 682, 2023
|
|
3DC8
 
 | |
5G3O
 
 | | Bacillus cereus formamidase (BceAmiF) inhibited with urea. | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, DI(HYDROXYETHYL)ETHER, FORMAMIDASE, ... | | Authors: | Gavira, J.A, Martinez-Rodriguez, S, Conejero-Muriel, M. | | Deposit date: | 2016-04-29 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A novel cysteine carbamoyl-switch is responsible for the inhibition of formamidase, a nitrilase superfamily member.
Arch.Biochem.Biophys., 662, 2019
|
|
5G3P
 
 | | Bacillus cereus formamidase (BceAmiF) acetylated at the active site. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gavira, J.A, Conejero-Muriel, M, Martinez-Rodriguez, S. | | Deposit date: | 2016-04-29 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A novel cysteine carbamoyl-switch is responsible for the inhibition of formamidase, a nitrilase superfamily member.
Arch.Biochem.Biophys., 662, 2019
|
|
6G5Z
 
 | |
6G60
 
 | |
3DIE
 
 | |
4LE9
 
 | | Crystal structure of a chimeric c-Src-SH3 domain | | Descriptor: | Proto-oncogene tyrosine-protein kinase Src, TRIETHYLENE GLYCOL | | Authors: | Camara-Artigas, A, Martinez-Rodriguez, S, Ortiz-Salmeron, E, Martin-Garcia, J.M. | | Deposit date: | 2013-06-25 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.344 Å) | | Cite: | 3D domain swapping in a chimeric c-Src SH3 domain takes place through two hinge loops.
J.Struct.Biol., 186, 2014
|
|
8QME
 
 | |
6YC5
 
 | |
5FQK
 
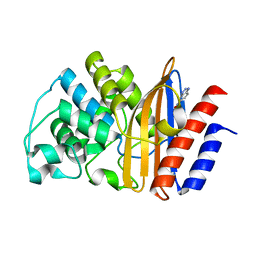 | | W229D and F290W mutant of the last common ancestor of Gram-negative bacteria (GNCA4) beta-lactamase class A bound to 5(6)-nitrobenzotriazole (TS-analog) | | Descriptor: | 6-NITROBENZOTRIAZOLE, GNCA4 LACTAMASE W229D AND F290W | | Authors: | Gavira, J.A, Risso, V.A, Martinez-Rodriguez, S, Sanchez-Ruiz, J.M. | | Deposit date: | 2015-12-11 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.767 Å) | | Cite: | De novo active sites for resurrected Precambrian enzymes.
Nat Commun, 8, 2017
|
|
7APP
 
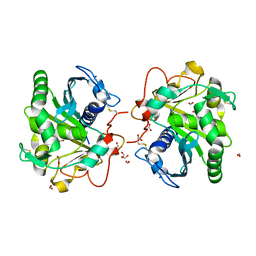 | | Structure of Lipase TL from capillary grown crystal in the presence of agarose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FORMIC ACID, Lipase, ... | | Authors: | Gavira, J.A, Martinez-Rodriguez, S, Fernande-Penas, R, Verdugo-Escamilla, C. | | Deposit date: | 2020-10-19 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Production of Cross-Linked Lipase Crystals at a Preparative Scale.
Cryst.Growth Des., 21, 2021
|
|
7APN
 
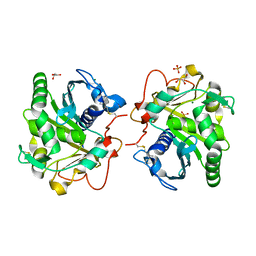 | | Structure of Lipase TL from bulk agarose grown crystal | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Lipase, ... | | Authors: | Gavira, J.A, Martinez-Rodriguez, S, Fernande-Penas, R, Verdugo-Escamilla, C. | | Deposit date: | 2020-10-19 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Production of Cross-Linked Lipase Crystals at a Preparative Scale.
Cryst.Growth Des., 21, 2021
|
|
7PTH
 
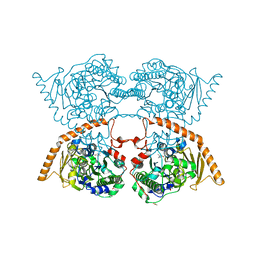 | |
7PTJ
 
 | |
5FQQ
 
 | | Last common ancestor of Gram-negative bacteria (GNCA4) beta-lactamase class A | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, DI(HYDROXYETHYL)ETHER, GNCA4 LACTAMASE | | Authors: | Gavira, J.A, Martinez-Rodriguez, S, Risso, V.A, Sanchez-Ruiz, J.M. | | Deposit date: | 2015-12-14 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | De novo active sites for resurrected Precambrian enzymes.
Nat Commun, 8, 2017
|
|
4UHU
 
 | | W229D mutant of the last common ancestor of Gram-negative bacteria (GNCA) beta-lactamase class A | | Descriptor: | ACETATE ION, FORMIC ACID, GNCA LACTAMASE W229D | | Authors: | Gavira, J.A, Risso, V.A, Martinez-Rodriguez, S, Sanchez-Ruiz, J.M. | | Deposit date: | 2015-03-25 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.305 Å) | | Cite: | De novo active sites for resurrected Precambrian enzymes.
Nat Commun, 8, 2017
|
|
4HX3
 
 | | Crystal structure of Streptomyces caespitosus sermetstatin in complex with S. caespitosus snapalysin | | Descriptor: | Extracellular small neutral protease, GLYCEROL, Neutral proteinase inhibitor ScNPI, ... | | Authors: | Trillo-Muyo, S, Martinez-Rodriguez, S, Arolas, J.L, Gomis-Ruth, F.X. | | Deposit date: | 2012-11-09 | | Release date: | 2012-12-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanism of action of a Janus-faced single-domain protein inhibitor simultaneously targeting two peptidase classes
CHEM SCI, 4, 2013
|
|
3EBA
 
 | | CAbHul6 FGLW mutant (humanized) in complex with human lysozyme | | Descriptor: | CAbHul6, Lysozyme C, SULFATE ION | | Authors: | Loris, R, Vincke, C, Saerens, D, Martinez-Rodriguez, S, Muyldermans, S, Conrath, K. | | Deposit date: | 2008-08-27 | | Release date: | 2008-12-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | General Strategy to Humanize a Camelid Single-domain Antibody and Identification of a Universal Humanized Nanobody Scaffold
J.Biol.Chem., 284, 2009
|
|
5FQJ
 
 | | W229D mutant of the last common ancestor of Gram-negative bacteria (GNCA) beta-lactamase bound to 5(6)-nitrobenzotriazole (TS-analog) | | Descriptor: | 6-NITROBENZOTRIAZOLE, GNCA LACTAMASE W229D | | Authors: | Gavira, J.A, Martinez-Rodriguez, S, Risso, V.A, Sanchez-Ruiz, J.M. | | Deposit date: | 2015-12-11 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.271 Å) | | Cite: | De novo active sites for resurrected Precambrian enzymes.
Nat Commun, 8, 2017
|
|
5FQI
 
 | | W229D and F290W mutant of the last common ancestor of Gram-negative bacteria (GNCA4) beta-lactamase class A | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gavira, J.A, Risso, V.A, Martinez-Rodriguez, S, Sanchez-Ruiz, J.M. | | Deposit date: | 2015-12-11 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | De novo active sites for resurrected Precambrian enzymes.
Nat Commun, 8, 2017
|
|
3EAK
 
 | | NbBCII10 humanized (FGLA mutant) | | Descriptor: | NbBCII10-FGLA, SULFATE ION | | Authors: | Vincke, C, Loris, R, Saerens, D, Martinez-Rodriguez, S, Muyldermans, S, Conrath, K. | | Deposit date: | 2008-08-26 | | Release date: | 2008-12-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | General strategy to humanize a camelid single-domain antibody and identification of a universal humanized nanobody scaffold.
J.Biol.Chem., 2008
|
|
6YBI
 
 | |
