8X7T
 
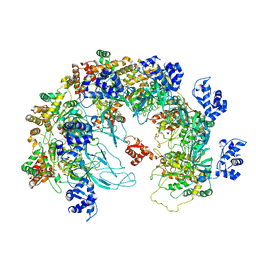 | | MCM in the Apo state. | | Descriptor: | mini-chromosome maintenance complex 3 | | Authors: | Ma, J, Yi, G, Ye, M, MacGregor-Chatwin, C, Sheng, Y, Lu, Y, Li, M, Gilbert, R.J.C, Zhang, P. | | Deposit date: | 2023-11-25 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | MCM in the Apo state
To Be Published
|
|
8X7U
 
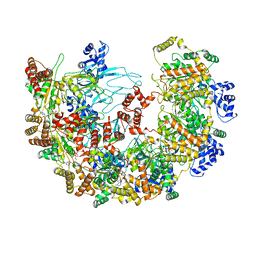 | | MCM in complex with dsDNA in presence of ATP. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, mini-chromosome maintenance complex 3 | | Authors: | Ma, J, Yi, G, Ye, M, MacGregor-Chatwin, C, Sheng, Y, Lu, Y, Li, M, Gilbert, R.J.C, Zhang, P. | | Deposit date: | 2023-11-25 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | MCM in complex with dsDNA in presence of ATP
To Be Published
|
|
1O5W
 
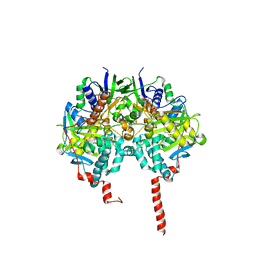 | | The structure basis of specific recognitions for substrates and inhibitors of rat monoamine oxidase A | | Descriptor: | Amine oxidase [flavin-containing] A, FLAVIN-ADENINE DINUCLEOTIDE, N-[3-(2,4-DICHLOROPHENOXY)PROPYL]-N-METHYL-N-PROP-2-YNYLAMINE | | Authors: | Ma, J, Yoshimura, M, Yamashita, E, Nakagawa, A, Ito, A, Tsukihara, T. | | Deposit date: | 2003-10-06 | | Release date: | 2004-04-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of rat monoamine oxidase a and its specific recognitions for substrates and inhibitors.
J.Mol.Biol., 338, 2004
|
|
6CSE
 
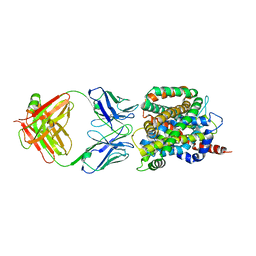 | | Crystal structure of sodium/alanine symporter AgcS with L-alanine bound | | Descriptor: | ALANINE, Monoclonal antibody FAB heavy chain, Monoclonal antibody FAB light chain, ... | | Authors: | Ma, J, Reyes, F.E, Gonen, T. | | Deposit date: | 2018-03-20 | | Release date: | 2019-01-30 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Structural basis for substrate binding and specificity of a sodium-alanine symporter AgcS.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6CSF
 
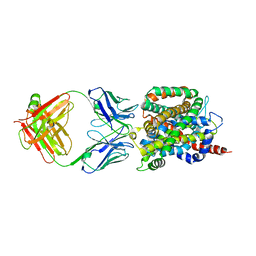 | | Crystal structure of sodium/alanine symporter AgcS with D-alanine bound | | Descriptor: | D-ALANINE, Monoclonal antibody FAB heavy chain, Monoclonal antibody FAB light chain, ... | | Authors: | Ma, J, Reyes, F.E, Gonen, T. | | Deposit date: | 2018-03-20 | | Release date: | 2019-01-30 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for substrate binding and specificity of a sodium-alanine symporter AgcS.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
2LR1
 
 | |
7EKA
 
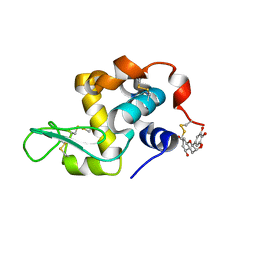 | | crystal structure of epigallocatechin binding with alpha-lactalbumin | | Descriptor: | 2-(3,4,5-TRIHYDROXY-PHENYL)-CHROMAN-3,5,7-TRIOL, Alpha-lactalbumin | | Authors: | Ma, J, Yao, Q, Chen, X, Zang, J. | | Deposit date: | 2021-04-05 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Weak Binding of Epigallocatechin to alpha-Lactalbumin Greatly Improves Its Stability and Uptake by Caco-2 Cells.
J.Agric.Food Chem., 69, 2021
|
|
3MIL
 
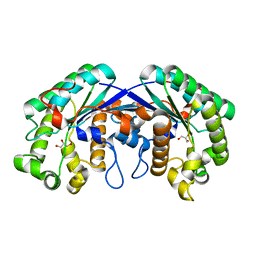 | | Crystal structure of isoamyl acetate-hydrolyzing esterase from Saccharomyces cerevisiae | | Descriptor: | GLYCEROL, Isoamyl acetate-hydrolyzing esterase | | Authors: | Ma, J, Lu, Q, Yuan, Y, Li, K, Ge, H, Go, Y, Niu, L, Teng, M. | | Deposit date: | 2010-04-11 | | Release date: | 2010-11-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of isoamyl acetate-hydrolyzing esterase from Saccharomyces cerevisiae reveals a novel active site architecture and the basis of substrate specificity
Proteins, 79, 2011
|
|
7VH3
 
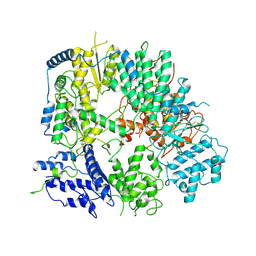 | |
7EXM
 
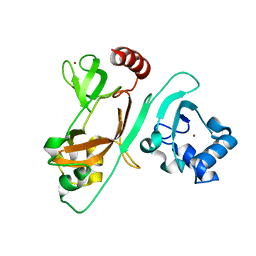 | | The N-terminal crystal structure of SARS-CoV-2 NSP2 | | Descriptor: | GLYCEROL, Non-structural protein 2, ZINC ION | | Authors: | Ma, J, Chen, Z. | | Deposit date: | 2021-05-27 | | Release date: | 2021-06-16 | | Last modified: | 2022-02-16 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure and Function of N-Terminal Zinc Finger Domain of SARS-CoV-2 NSP2.
Virol Sin, 36, 2021
|
|
5GQQ
 
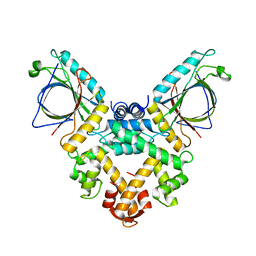 | | Structure of ALG-2/HEBP2 Complex | | Descriptor: | CALCIUM ION, CHLORIDE ION, Heme-binding protein 2, ... | | Authors: | Liu, X, Ma, J, Zhang, H, Feng, Y. | | Deposit date: | 2016-08-08 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Functional Study of Apoptosis-linked Gene-2Heme-binding Protein 2 Interactions in HIV-1 Production.
J. Biol. Chem., 291, 2016
|
|
4FHZ
 
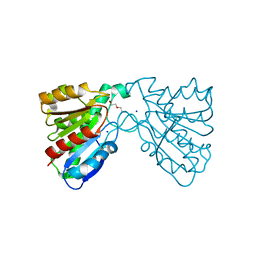 | | Crystal structure of a carboxyl esterase at 2.0 angstrom resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, Phospholipase/Carboxylesterase, SODIUM ION | | Authors: | Wu, L, Ma, J, Zhou, J, Yu, H. | | Deposit date: | 2012-06-07 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Enhanced enantioselectivity of a carboxyl esterase from Rhodobacter sphaeroides by directed evolution.
Appl.Microbiol.Biotechnol., 97, 2013
|
|
4FTW
 
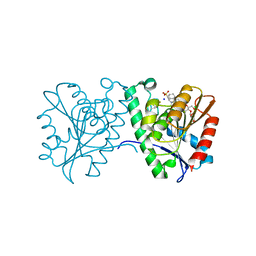 | | Crystal structure of a carboxyl esterase N110C/L145H at 2.3 angstrom resolution | | Descriptor: | 3-CYCLOHEXYLPROPYL 4-O-ALPHA-D-GLUCOPYRANOSYL-BETA-D-GLUCOPYRANOSIDE, CHLORIDE ION, PIPERAZINE-N,N'-BIS(2-ETHANESULFONIC ACID), ... | | Authors: | Wu, L, Ma, J, Zhou, J, Yu, H. | | Deposit date: | 2012-06-28 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enhanced enantioselectivity of a carboxyl esterase from Rhodobacter sphaeroides by directed evolution.
Appl.Microbiol.Biotechnol., 97, 2013
|
|
3BLI
 
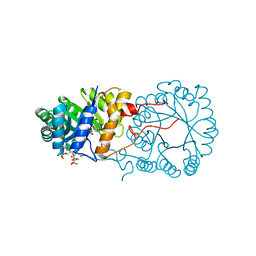 | |
3BLF
 
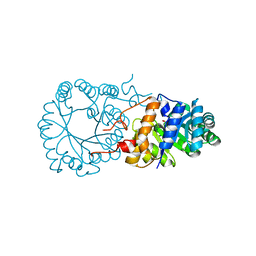 | |
3BLE
 
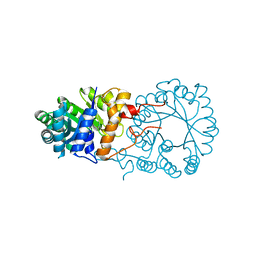 | |
7E7D
 
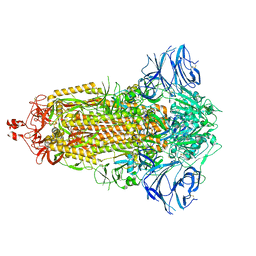 | |
7E7B
 
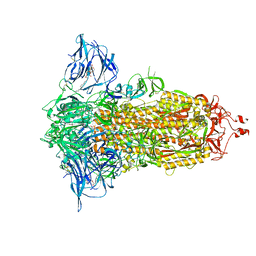 | | Cryo-EM structure of the SARS-CoV-2 furin site mutant S-Trimer from a subunit vaccine candidate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-hydroxyethyl 2-deoxy-3,5-bis-O-(2-hydroxyethyl)-6-O-(2-{[(9E)-octadec-9-enoyl]oxy}ethyl)-alpha-L-xylo-hexofuranoside, ... | | Authors: | Zheng, S, Ma, J. | | Deposit date: | 2021-02-25 | | Release date: | 2021-03-24 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Cryo-EM structure of S-Trimer, a subunit vaccine candidate for COVID-19.
J.Virol., 95, 2021
|
|
6UZ1
 
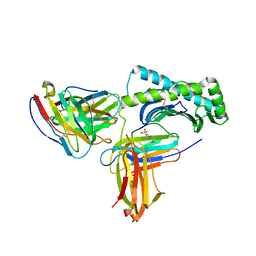 | |
7RRG
 
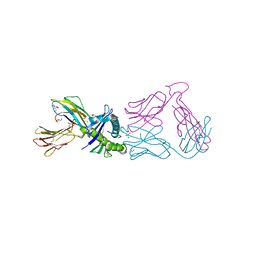 | |
4F33
 
 | | Crystal Structure of therapeutic antibody MORAb-009 | | Descriptor: | MORAb-009 FAB heavy chain, MORAb-009 FAB light chain, TETRAETHYLENE GLYCOL | | Authors: | Xia, D, Ma, J, Tang, W.K, Esser, L. | | Deposit date: | 2012-05-08 | | Release date: | 2012-07-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | Recognition of mesothelin by the therapeutic antibody MORAb-009: structural and mechanistic insights.
J.Biol.Chem., 287, 2012
|
|
4F3F
 
 | | Crystal Structure of Msln7-64 MORAb-009 FAB complex | | Descriptor: | MORAb-009 Fab heavy chain, MORAb-009 Fab light chain, Mesothelin | | Authors: | Xia, D, Pastan, I, Ma, J, Tang, W.K, Esser, L. | | Deposit date: | 2012-05-09 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Recognition of mesothelin by the therapeutic antibody MORAb-009: structural and mechanistic insights.
J.Biol.Chem., 287, 2012
|
|
6JCN
 
 | | Yeast dehydrodolichyl diphosphate synthase complex subunit NUS1 | | Descriptor: | Dehydrodolichyl diphosphate synthase complex subunit NUS1, SULFATE ION | | Authors: | Ko, T.-P, Ma, J, Liu, W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2019-01-29 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structural insights to heterodimeric cis-prenyltransferases through yeast dehydrodolichyl diphosphate synthase subunit Nus1.
Biochem.Biophys.Res.Commun., 515, 2019
|
|
7VGQ
 
 | | Cryo-EM structure of Machupo virus polymerase L in complex with matrix protein Z | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,RING finger protein Z, RNA-directed RNA polymerase L, ZINC ION | | Authors: | Zhang, X, Ma, J, Zhang, S. | | Deposit date: | 2021-09-18 | | Release date: | 2021-09-29 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of Machupo virus polymerase in complex with matrix protein Z.
Nat Commun, 12, 2021
|
|
7VH2
 
 | |
