1F33
 
 | |
1F30
 
 | | THE STRUCTURAL BASIS FOR DNA PROTECTION BY E. COLI DPS PROTEIN | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA PROTECTION DURING STARVATION PROTEIN, ZINC ION | | Authors: | Luo, J, Liu, D, White, M.A, Fox, R.O. | | Deposit date: | 2000-05-31 | | Release date: | 2003-06-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The Structural Basis for DNA Protection by E. coli Dps Protein
To be Published
|
|
4QHU
 
 | | Crystal Structure of IL-17A/Fab6785 complex | | Descriptor: | CHLORIDE ION, Fab6785 heavy chain, Fab6785 light chain, ... | | Authors: | Luo, J, Gilliland, G.L, Malia, T, Obmolova, G, Teplyakov, A. | | Deposit date: | 2014-05-29 | | Release date: | 2015-08-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | IL-17A Asymmetry in a Complex with a Neutralizing Antibody Fab6785 Reveals a Potential Mechanism of Receptor Differentiation
To be Published
|
|
3HMX
 
 | | Crystal structure of ustekinumab FAB/IL-12 complex | | Descriptor: | Interleukin-12 subunit alpha, Interleukin-12 subunit beta, USTEKINUMAB FAB HEAVY CHAIN, ... | | Authors: | Luo, J. | | Deposit date: | 2009-05-29 | | Release date: | 2010-06-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the dual recognition of IL-12 and IL-23 by ustekinumab.
J.Mol.Biol., 402, 2010
|
|
3HMW
 
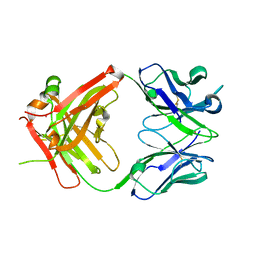 | | Crystal structure of ustekinumab FAB | | Descriptor: | CADMIUM ION, USTEKINUMAB FAB HEAVY CHAIN, USTEKINUMAB FAB LIGHT CHAIN | | Authors: | Luo, J. | | Deposit date: | 2009-05-29 | | Release date: | 2010-06-09 | | Last modified: | 2015-07-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the dual recognition of IL-12 and IL-23 by ustekinumab.
J.Mol.Biol., 402, 2010
|
|
3I84
 
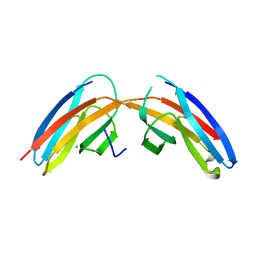 | |
3I85
 
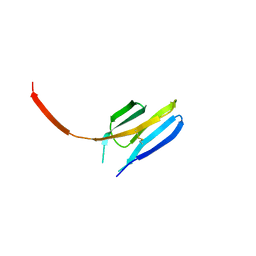 | |
3QQ9
 
 | | Crystal structure of FAB fragment of anti-human RSV (RESPIRATORY SYNCYTIAL VIRUS) F Protein MAB 101F | | Descriptor: | 101F HEAVY CHAIN, 101F LIGHT CHAIN, SULFATE ION | | Authors: | Luo, J, Tsui, P, Spurlino, J, Lewansowski, F, Heavner, G.A, Del Vecchio, F. | | Deposit date: | 2011-02-15 | | Release date: | 2011-08-10 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal Structure of Fab 101F
To be Published
|
|
3QRG
 
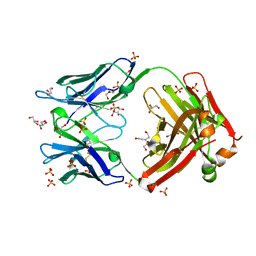 | | Crystal structure of antiRSVF Fab B21m | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Fab heavy chain Fd fragment, Fab light chain, ... | | Authors: | Luo, J, Tsui, P, Lewandowski, F, Spurlino, J, Vecchio, F.D. | | Deposit date: | 2011-02-17 | | Release date: | 2011-08-10 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Crystal structure anti-RSV Fab B21m
To be Published
|
|
4M6A
 
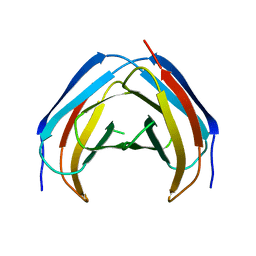 | | N-Terminal beta-Strand Swapping in a Consensus Derived Alternative Scaffold Driven by Stabilizing Hydrophobic Interactions | | Descriptor: | Tencon | | Authors: | Luo, J, Teplyakov, A, Obmolova, G, Malia, T.J, Chan, W, Jocobs, S.A, O'neil, K.T, Gilliland, G.L. | | Deposit date: | 2013-08-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | N-terminal beta-strand swapping in a consensus-derived alternative scaffold driven by stabilizing hydrophobic interactions.
Proteins, 82, 2014
|
|
3NAB
 
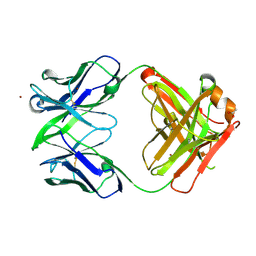 | | Crystal Structure of fab15 Mut6 | | Descriptor: | ACETATE ION, Fab15 Mut6 heavy chain, Fab15 Mut6 light chain, ... | | Authors: | Luo, J. | | Deposit date: | 2010-06-01 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Co-evolution of antibody stability and Vk CDR-L3 canonical structure
To be Published
|
|
3NAC
 
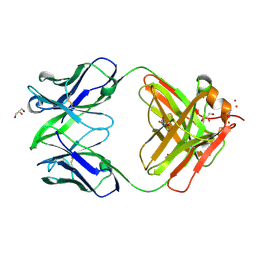 | | Crystal structure of Fab15 Mut7 | | Descriptor: | ACETATE ION, Fab15 Mut7 heavy chain, Fab15 Mut7 light chain, ... | | Authors: | Luo, J. | | Deposit date: | 2010-06-01 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Co-evolution of antibody stability and Vk CDR-L3 canonical structure
To be Published
|
|
3NCJ
 
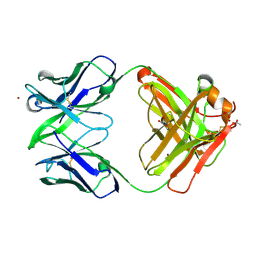 | | Crystal structure of Fab15 Mut8 | | Descriptor: | ACETATE ION, Fab15 Mut8 heavy chain, Fab15 Mut8 light chain, ... | | Authors: | Luo, J, Feng, Y, Gilliland, G.L. | | Deposit date: | 2010-06-04 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Co-evolution of antibody stability and Vk CDR-L3 canonical structure
To be Published
|
|
3NAA
 
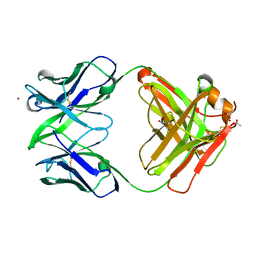 | | Crystal structure of Fab15 Mut5 | | Descriptor: | ACETATE ION, Fab15 Mut5 heavy chain, Fab15 Mut5 light chain, ... | | Authors: | Luo, J, Feng, Y, Gilliland, G.L. | | Deposit date: | 2010-06-01 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Co-evolution of antibody stability and Vk CDR-L3 canonical structure
To be Published
|
|
3NA9
 
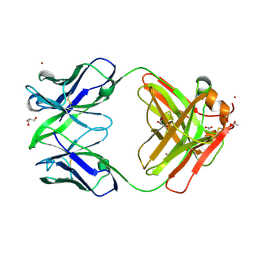 | | Crystal structure of Fab15 | | Descriptor: | ACETATE ION, Fab15 heavy chain, Fab15 light chain, ... | | Authors: | Luo, J. | | Deposit date: | 2010-06-01 | | Release date: | 2010-08-18 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Co-evolution of antibody stability and Vk CDR-L3 canonical structure
To be Published
|
|
7V1N
 
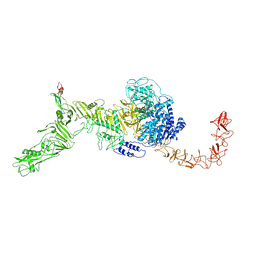 | | Structure of the Clade 2 C. difficile TcdB in complex with its receptor TFPI | | Descriptor: | Isoform Beta of Tissue factor pathway inhibitor, Toxin B | | Authors: | Luo, J, Yang, Q, Zhang, X, Zhang, Y, Wan, L, Li, Y, Tao, L. | | Deposit date: | 2021-08-05 | | Release date: | 2022-02-23 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | TFPI is a colonic crypt receptor for TcdB from hypervirulent clade 2 C. difficile.
Cell, 185, 2022
|
|
3QPX
 
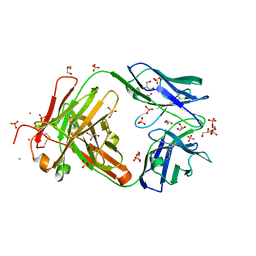 | | Crystal structure of Fab C2507 | | Descriptor: | CADMIUM ION, CHLORIDE ION, Fab C2507 heavy chain, ... | | Authors: | Luo, J, Teplyakov, A, Obmolova, O, Malia, T, Gilliland, G.L. | | Deposit date: | 2011-02-14 | | Release date: | 2011-08-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Fab C2507
To be Published
|
|
3QPQ
 
 | | Crystal structure of ANTI-TLR3 antibody C1068 FAB | | Descriptor: | C1068 heavy chain, C1068 light chain, GLYCEROL, ... | | Authors: | Luo, J, Obmolova, G, Teplyakov, A, Gilliland, G.L. | | Deposit date: | 2011-02-14 | | Release date: | 2011-08-10 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Fab C1068
To be Published
|
|
1JRE
 
 | | DNA PROTECTION AND BINDING BY E. COLI DPS PROTEIN | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CADMIUM ION, DNA PROTECTION DURING STARVATION PROTEIN | | Authors: | Luo, J, Liu, D, White, M.A, Fox, R.O. | | Deposit date: | 2001-08-13 | | Release date: | 2003-06-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | DNA Protection and Binding by E. Coli Dps Protein
To be Published
|
|
1JTS
 
 | |
1L8H
 
 | | DNA PROTECTION AND BINDING BY E. COLI DPS PROTEIN | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA PROTECTION DURING STARVATION PROTEIN, POTASSIUM ION | | Authors: | Luo, J, Liu, D, White, M.A, Fox, R.O. | | Deposit date: | 2002-03-20 | | Release date: | 2003-06-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | DNA Protection and Binding by E. Coli Dps Protein
To be Published
|
|
1L8I
 
 | | Dna Protection and Binding by E. Coli DPS Protein | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA PROTECTION DURING STARVATION PROTEIN, POTASSIUM ION | | Authors: | Luo, J, Liu, D, White, M.A, Fox, R.O. | | Deposit date: | 2002-03-20 | | Release date: | 2003-06-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | DNA Protection and Binding by E. Coli Dps Protein
To be Published
|
|
1R8T
 
 | | Solution structures of high affinity miniprotein ligands to Streptavidin | | Descriptor: | MP1 | | Authors: | Luo, J, Mukherjee, M, Fan, X, Yang, H, Liu, D, Khan, R, White, M, Fox, R.O. | | Deposit date: | 2003-10-28 | | Release date: | 2005-02-15 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure-based design of high affinity miniprotein ligands
To be Published
|
|
3ULS
 
 | | Crystal structure of Fab12 | | Descriptor: | Fab12 heavy chain, Fab12 light chain | | Authors: | Luo, J, Gilliland, G.L, Obmolova, O, Malia, T, Teplyakov, A. | | Deposit date: | 2011-11-11 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Lateral Clustering of TLR3:dsRNA Signaling Units Revealed by TLR3ecd:3Fabs Quaternary Structure.
J.Mol.Biol., 421, 2012
|
|
3ULU
 
 | | Structure of quaternary complex of human TLR3ecd with three Fabs (Form1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab1068 heavy chain, ... | | Authors: | Luo, J, Gilliland, G.L, Obmolova, O, Malia, T, Teplyakov, A. | | Deposit date: | 2011-11-11 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | Lateral Clustering of TLR3:dsRNA Signaling Units Revealed by TLR3ecd:3Fabs Quaternary Structure.
J.Mol.Biol., 421, 2012
|
|
