1K0F
 
 | | Crystal structure of Zn(II)-free T. pallidum TroA | | Descriptor: | Periplasmic zinc-binding protein troA | | Authors: | Lee, Y.H, Dorwart, M.R, Hazlett, K.R, Deka, R.K, Norgard, M.V, Radolf, J.D, Hasemann, C.A. | | Deposit date: | 2001-09-19 | | Release date: | 2002-04-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of Zn(II)-free Treponema pallidum TroA, a periplasmic metal-binding protein, reveals a closed conformation.
J.Bacteriol., 184, 2002
|
|
1K6M
 
 | | Crystal Structure of Human Liver 6-Phosphofructo-2-Kinase/Fructose-2,6-Bisphosphatase | | Descriptor: | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2-phosphatase, PHOSPHATE ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Lee, Y.H, Li, Y, Uyeda, K, Hasemann, C.A. | | Deposit date: | 2001-10-16 | | Release date: | 2002-12-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Tissue-specific structure/function differentiation of the liver isoform of 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase.
J.Biol.Chem., 278, 2003
|
|
1TOA
 
 | | PERIPLASMIC ZINC BINDING PROTEIN TROA FROM TREPONEMA PALLIDUM | | Descriptor: | GLYCEROL, PROTEIN (PERIPLASMIC BINDING PROTEIN TROA), ZINC ION | | Authors: | Lee, Y.H, Deka, R.K, Norgard, M.V, Radolf, J.D, Hasemann, C.A. | | Deposit date: | 1999-03-03 | | Release date: | 1999-07-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Treponema pallidum TroA is a periplasmic zinc-binding protein with a helical backbone.
Nat.Struct.Biol., 6, 1999
|
|
1C80
 
 | | REGULATORY COMPLEX OF FRUCTOSE-2,6-BISPHOSPHATASE | | Descriptor: | FRUCTOSE-2,6-BISPHOSPHATASE, GUANOSINE-5'-TRIPHOSPHATE, PHOSPHATE ION | | Authors: | Lee, Y.H, Olson, T.W, McClard, R.W, Witte, J.F, McFarlan, S.C, Banaszak, L.J, Levitt, D.G, Lange, A.J. | | Deposit date: | 2000-04-03 | | Release date: | 2003-06-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reaction Mechanism of Fructose-2,6-bisphosphatase Suggested by the Crystal Structures of a pseudo-Michaelis complex and Metabolite Complexes
To be Published
|
|
1C7Z
 
 | | REGULATORY COMPLEX OF FRUCTOSE-2,6-BISPHOSPHATASE | | Descriptor: | FRUCTOSE-2,6-BISPHOSPHATASE, GLYCERALDEHYDE-3-PHOSPHATE, PHOSPHATE ION | | Authors: | Lee, Y.H, Olson, T.W, McClard, R.W, Witte, J.F, McFarlan, S.C, Banaszak, L.J, Levitt, D.G, Lange, A.J. | | Deposit date: | 2000-04-03 | | Release date: | 2003-06-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Reaction Mechanism of Fructose-2,6-bisphosphatase Suggested by the Crystal Structures of a pseudo-Michaelis complex and Metabolite Complexes
To be Published
|
|
1C81
 
 | | MICHAELIS COMPLEX OF FRUCTOSE-2,6-BISPHOSPHATASE | | Descriptor: | 2,5-anhydro-1-deoxy-1-phosphono-6-O-phosphono-D-glucitol, FRUCTOSE-2,6-BISPHOSPHATASE | | Authors: | Lee, Y.H, Olson, T.W, McClard, R.W, Witte, J.F, McFarlan, S.C, Banaszak, L.J, Levitt, D.G, Lange, A.J. | | Deposit date: | 2000-04-03 | | Release date: | 2003-06-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Reaction Mechanism of Fructose-2,6-bisphosphatase Suggested by the Crystal Structures of a pseudo-Michaelis complex and Metabolite Complexes
To be Published
|
|
2DWO
 
 | | PFKFB3 in complex with ADP and PEP | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Lee, Y.H. | | Deposit date: | 2006-08-16 | | Release date: | 2007-07-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A Direct Substrate-Substrate Interaction Found in the Kinase Domain of the Bifunctional Enzyme, 6-Phosphofructo-2-kinase/Fructose-2,6-bisphosphatase
J.Mol.Biol., 370, 2007
|
|
2F7N
 
 | | Structure of D. radiodurans Dps-1 | | Descriptor: | COBALT (II) ION, DNA-binding stress response protein, Dps family, ... | | Authors: | Lee, Y.H, Kim, S.G, Bhattacharyya, G, Grove, A. | | Deposit date: | 2005-12-01 | | Release date: | 2006-11-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Dps-1, a functionally distinct Dps protein from Deinococcus radiodurans.
J.Mol.Biol., 361, 2006
|
|
2DWP
 
 | | A pseudo substrate complex of 6-phosphofructo-2-kinase of PFKFB | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3, MAGNESIUM ION, ... | | Authors: | Lee, Y.H. | | Deposit date: | 2006-08-16 | | Release date: | 2007-07-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Direct Substrate-Substrate Interaction Found in the Kinase Domain of the Bifunctional Enzyme, 6-Phosphofructo-2-kinase/Fructose-2,6-bisphosphatase
J.Mol.Biol., 370, 2007
|
|
2A3M
 
 | | Structure of Desulfovibrio desulfuricans G20 tetraheme cytochrome (oxidized form) | | Descriptor: | COG3005: Nitrate/TMAO reductases, membrane-bound tetraheme cytochrome c subunit, HEME C | | Authors: | Pattarkine, M.V, Tanner, J.J, Bottoms, C.A, Lee, Y.H, Wall, J.D. | | Deposit date: | 2005-06-25 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Desulfovibrio desulfuricans G20 Tetraheme Cytochrome Structure at 1.5A and Cytochrome Interaction with Metal Complexes
J.Mol.Biol., 358, 2006
|
|
3AXG
 
 | | Structure of 6-aminohexanoate-oligomer hydrolase | | Descriptor: | Endotype 6-aminohexanoat-oligomer hydrolase, SODIUM ION | | Authors: | Negoro, S, Shibata, N, Tanaka, Y, Yasuhira, K, Shibata, H, Hashimoto, H, Lee, Y.H, Ohshima, S, Santa, R, Mochiji, K, Goto, Y, Ikegami, T, Nagai, K, Kato, D, Takeo, M, Higuchi, Y. | | Deposit date: | 2011-04-04 | | Release date: | 2011-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of nylon hydrolase and mechanism of nylon-6 hydrolysis
J.Biol.Chem., 287, 2012
|
|
1XVJ
 
 | | Crystal Structure Of Rat alpha-Parvalbumin D94S/G98E Mutant | | Descriptor: | CALCIUM ION, Parvalbumin alpha | | Authors: | Tanner, J.J, Agah, S, Lee, Y.H, Henzl, M.T. | | Deposit date: | 2004-10-28 | | Release date: | 2005-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the D94S/G98E Variant of Rat alpha-Parvalbumin. An Explanation for the Reduced Divalent Ion Affinity.
Biochemistry, 44, 2005
|
|
2AXN
 
 | | Crystal structure of the human inducible form 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (6PF-2-K/Fru- 2,6-P2ASE brain/placenta-type isozyme) (iPFK-2) [Includes: 6- phosphofructo-2-kinase (EC 2.7.1.105); Fructose-2,6-bisphosphatase (EC 3.1.3.46)], ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kim, S.G, Manes, N.P, El-Maghrabi, M.R, Lee, Y.H. | | Deposit date: | 2005-09-05 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the hypoxia-inducible form of 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase (PFKFB3): a possible new target for cancer therapy.
J.Biol.Chem., 281, 2006
|
|
5AZ2
 
 | | Crystal structure of the Fab fragment of 9E5, a murine monoclonal antibody specific for human epiregulin | | Descriptor: | anti-human epiregulin antibody 9E5 Fab heavy chain, anti-human epiregulin antibody 9E5 Fab light chain | | Authors: | Kado, Y, Mizohata, E, Nagatoishi, S, Iijima, M, Shinoda, K, Miyafusa, T, Nakayama, T, Yoshizumi, T, Sugiyama, A, Kawamura, T, Lee, Y.H, Matsumura, H, Doi, H, Fujitani, H, Kodama, T, Shibasaki, Y, Tsumoto, K, Inoue, T. | | Deposit date: | 2015-09-16 | | Release date: | 2015-12-09 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Epiregulin Recognition Mechanisms by Anti-epiregulin Antibody 9E5: STRUCTURAL, FUNCTIONAL, AND MOLECULAR DYNAMICS SIMULATION ANALYSES
J.Biol.Chem., 291, 2016
|
|
5E8D
 
 | | Crystal structure of human epiregulin in complex with the Fab fragment of murine monoclonal antibody 9E5 | | Descriptor: | CHLORIDE ION, GLYCEROL, Proepiregulin, ... | | Authors: | Kado, Y, Mizohata, E, Nagatoishi, S, Iijima, M, Shinoda, K, Miyafusa, T, Nakayama, T, Yoshizumi, T, Sugiyama, A, Kawamura, T, Lee, Y.H, Matsumura, H, Doi, H, Fujitani, H, Kodama, T, Shibasaki, Y, Tsumoto, K, Inoue, T. | | Deposit date: | 2015-10-14 | | Release date: | 2015-12-09 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Epiregulin Recognition Mechanisms by Anti-epiregulin Antibody 9E5: STRUCTURAL, FUNCTIONAL, AND MOLECULAR DYNAMICS SIMULATION ANALYSES
J.Biol.Chem., 291, 2016
|
|
2I1V
 
 | | Crystal structure of PFKFB3 in complex with ADP and Fructose-2,6-bisphosphate | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, 6-O-phosphono-beta-D-fructofuranose, 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3, ... | | Authors: | Kim, S.G, El-Maghrabi, M.R, Lee, Y.H. | | Deposit date: | 2006-08-15 | | Release date: | 2007-07-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Direct Substrate-Substrate Interaction Found in the Kinase Domain of the Bifunctional Enzyme, 6-Phosphofructo-2-kinase/Fructose-2,6-bisphosphatase
J.Mol.Biol., 370, 2007
|
|
3QPU
 
 | | PFKFB3 in complex with PPi | | Descriptor: | 1,2-ETHANEDIOL, 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3, PYROPHOSPHATE 2-, ... | | Authors: | Cavalier, M.C, Kim, S.G, Neau, D, Lee, Y.H. | | Deposit date: | 2011-02-14 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis of the fructose-2,6-bisphosphatase reaction of PFKFB3: Transition state and the C-terminal function.
Proteins, 80, 2012
|
|
3QPV
 
 | | PFKFB3 trapped in a phospho-enzyme intermediate state | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, 6-O-phosphono-beta-D-fructofuranose, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3, ... | | Authors: | Cavalier, M.C, Kim, S.G, Neau, D, Lee, Y.H. | | Deposit date: | 2011-02-14 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis of the fructose-2,6-bisphosphatase reaction of PFKFB3: Transition state and the C-terminal function.
Proteins, 80, 2012
|
|
3QPW
 
 | | PFKFB3 in complex with Aluminum Tetrafluoride | | Descriptor: | 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Cavalier, M.C, Kim, S.G, Neau, D, Lee, Y.H. | | Deposit date: | 2011-02-14 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Molecular basis of the fructose-2,6-bisphosphatase reaction of PFKFB3: Transition state and the C-terminal function.
Proteins, 80, 2012
|
|
1TIP
 
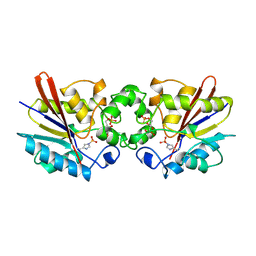 | | THE BISPHOSPHATASE DOMAIN OF THE BIFUNCTIONAL RAT LIVER 6-PHOSPHOFRUCTO-2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, PHOSPHOENZYME INTERMEDIATE OF FRU-2,6-BISPHOSPHATASE | | Authors: | Lee, Y.-H, Olson, T.W, Ogata, C.M, Levitt, D.G, Banaszak, L.J, Lange, A.J. | | Deposit date: | 1997-05-28 | | Release date: | 1998-01-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a trapped phosphoenzyme during a catalytic reaction.
Nat.Struct.Biol., 4, 1997
|
|
1FBT
 
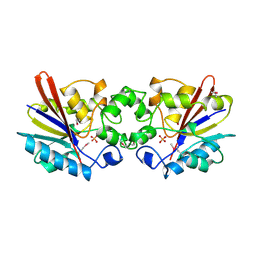 | | THE BISPHOSPHATASE DOMAIN OF THE BIFUNCTIONAL RAT LIVER 6-PHOSPHOFRUCTO-2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE | | Descriptor: | FRUCTOSE-2,6-BISPHOSPHATASE, PHOSPHATE ION | | Authors: | Lee, Y.-H, Ogata, C, Pflugrath, J.W, Levitt, D.G, Sarma, R, Banaszak, L.J, Pilkis, S.J. | | Deposit date: | 1996-03-08 | | Release date: | 1997-07-23 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the rat liver fructose-2,6-bisphosphatase based on selenomethionine multiwavelength anomalous dispersion phases.
Biochemistry, 35, 1996
|
|
4O8A
 
 | |
1S3P
 
 | |
7VOC
 
 | | The crystal structure of a Radical SAM Enzyme BlsE involved in the Biosynthesis of Blasticidin S | | Descriptor: | (2~{S},3~{S},4~{S},5~{R},6~{R})-6-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4,5-tris(oxidanyl)oxane-2-carboxylic acid, Cytosylglucuronate decarboxylase, GLYCEROL, ... | | Authors: | Hou, X.L, Zhou, J.H. | | Deposit date: | 2021-10-13 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.62005424 Å) | | Cite: | Radical S -Adenosyl Methionine Enzyme BlsE Catalyzes a Radical-Mediated 1,2-Diol Dehydration during the Biosynthesis of Blasticidin S.
J.Am.Chem.Soc., 144, 2022
|
|
7VOB
 
 | |
