8BB2
 
 | | Structure of human WDR5 and pVHL:ElonginC:ElonginB bound to PROTAC with PEG linker (conformation #2) | | Descriptor: | 1,2-ETHANEDIOL, Elongin-B, Elongin-C, ... | | Authors: | Kraemer, A, Doelle, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-10-12 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of human WDR5 and pVHL:ElonginC:ElonginB bound to PROTAC with PEG linker (conformation #2)
To Be Published
|
|
8BB3
 
 | | Structure of human WDR5 and pVHL:ElonginC:ElonginB bound to PROTAC with PEG linker (conformation #1) | | Descriptor: | 1,2-ETHANEDIOL, Elongin-B, Elongin-C, ... | | Authors: | Kraemer, A, Doelle, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-10-12 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of human WDR5 and pVHL:ElonginC:ElonginB bound to PROTAC with PEG linker (conformation #1)
To Be Published
|
|
8BEM
 
 | |
8BFM
 
 | | Crystal structure of human calmodulin-dependent protein kinase 1D (CAMK1D) in complex with FZ331 | | Descriptor: | Calcium/calmodulin-dependent protein kinase type 1D, SULFATE ION, pyrazolo[5,1-a]phthalazin-6-amine | | Authors: | Kraemer, A, Zhu, W.F, Hernandez-Olmos, V, Proschak, E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-10-26 | | Release date: | 2022-11-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of human WDR5 and pVHL:ElonginC:ElonginB bound to PROTAC with PEG linker (conformation #2)
To Be Published
|
|
7QGJ
 
 | | Apo structure of BIR2 Domain of BIRC2 | | Descriptor: | 1,2-ETHANEDIOL, Baculoviral IAP repeat-containing protein 2, ZINC ION | | Authors: | Kraemer, A, Farges, F, Schwalm, M.P, Saxena, K, Preuss, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-12-08 | | Release date: | 2022-02-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Apo structure BIR2 Domain of BIRC2
To Be Published
|
|
8BFS
 
 | | Crystal structure of human calmodulin-dependent protein kinase 1D (CAMK1D) in complex with FZ326 | | Descriptor: | 1,2-ETHANEDIOL, 3~{H}-pyrrolo[2,3-c]isoquinolin-5-amine, Calcium/calmodulin-dependent protein kinase type 1D, ... | | Authors: | Kraemer, A, Zhu, W.F, Hernandez-Olmos, V, Proschak, E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-10-26 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of human calmodulin-dependent protein kinase 1D (CAMK1D) in complex with FZ326
To Be Published
|
|
9FYF
 
 | | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with TR06772818 | | Descriptor: | Casein kinase II subunit alpha, SULFATE ION, ~{N}-[5-[[3-cyano-7-(cyclopropylamino)pyrazolo[1,5-a]pyrimidin-5-yl]amino]-4-fluoranyl-2-[(3~{S})-3-(methylamino)piperidin-1-yl]phenyl]propanamide | | Authors: | Kraemer, A, Ong, H.W, Yang, X, Brown, J.W, Chang, E, Willson, T, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-07-03 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with TR06772818
To Be Published
|
|
9FLQ
 
 | | Crystal structure of human Haspin (GSG2) kinase bound to MU1959 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, NICKEL (II) ION, ... | | Authors: | Kraemer, A, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-06-05 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of human Haspin (GSG2) kinase bound to MU1959
To Be Published
|
|
9I2Y
 
 | | Human DHODH in Complex with QC6352 | | Descriptor: | 3-[({(1R)-6-[methyl(phenyl)amino]-1,2,3,4-tetrahydronaphthalen-1-yl}methyl)amino]pyridine-4-carboxylic acid, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Kraemer, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2025-01-22 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Human DHODH in Complex with QC6352
To Be Published
|
|
9EZ3
 
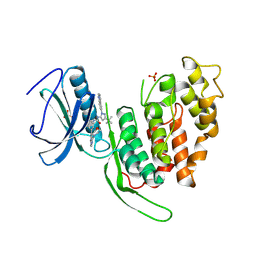 | | Crystal structure of human CLK3 bound to RN129 | | Descriptor: | 1-(5-~{tert}-butyl-2-quinolin-6-yl-pyrazol-3-yl)-3-[3-(4-morpholin-4-yl-7~{H}-pyrrolo[2,3-d]pyrimidin-5-yl)phenyl]urea, Dual specificity protein kinase CLK3, SULFATE ION | | Authors: | Kraemer, A, Raig, N, Knapp, S. | | Deposit date: | 2024-04-10 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of human CLK3 bound to RN129
To Be Published
|
|
8QWZ
 
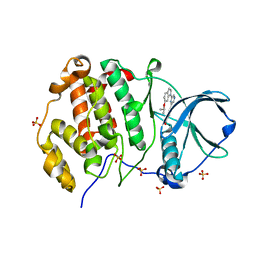 | | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with 4-(6-(6-isopropoxy-1H-indol-1-yl)pyrazin-2-yl)benzoic acid | | Descriptor: | 4-[6-(6-propan-2-yloxyindol-1-yl)pyrazin-2-yl]benzoic acid, Casein kinase II subunit alpha, SULFATE ION | | Authors: | Kraemer, A, Galal, K, Willson, T, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-20 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with 4-(6-(6-isopropoxy-1H-indol-1-yl)pyrazin-2-yl)benzoic acid
To Be Published
|
|
8QWY
 
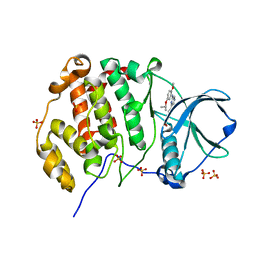 | | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with 4-(6-((5-isopropoxy-2-methoxyphenyl)amino)pyrazin-2-yl)benzoic acid | | Descriptor: | 4-[6-[(2-methoxy-5-propan-2-yloxy-phenyl)amino]pyrazin-2-yl]benzoic acid, Casein kinase II subunit alpha, SULFATE ION | | Authors: | Kraemer, A, Galal, K, Willson, T, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-20 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with 4-(6-(6-isopropoxy-1H-indol-1-yl)pyrazin-2-yl)benzoic acid
To Be Published
|
|
6TCX
 
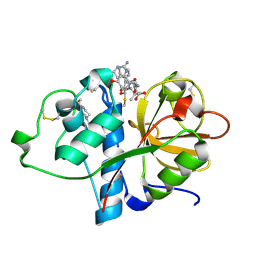 | | Papain bound to a natural cysteine protease inhibitor from Streptomyces mobaraensis | | Descriptor: | (2~{R})-2-[[(1~{S})-1-[(6~{S})-2-azanyl-1,4,5,6-tetrahydropyrimidin-6-yl]-2-[[(2~{S})-3-methyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanyl-3-phenyl-propan-2-yl]amino]butan-2-yl]amino]-2-oxidanylidene-ethyl]carbamoylamino]-3-(4-hydroxyphenyl)propanoic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Papain | | Authors: | Kraemer, A, Juettner, N.E, Fuchsbauer, H.-L, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-06 | | Release date: | 2019-12-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Decoding the Papain Inhibitor from Streptomyces mobaraensis as Being Hydroxylated Chymostatin Derivatives: Purification, Structure Analysis, and Putative Biosynthetic Pathway.
J.Nat.Prod., 83, 2020
|
|
9QBA
 
 | | Human TRIM21 PRYSPRY domain in complex with AL236 | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase TRIM21, ~{N}-(cyclohexylmethyl)-4-(4-fluoranyl-2-methylsulfanyl-phenyl)-2-methylsulfonyl-benzamide | | Authors: | Kim, Y, Lucic, A, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2025-03-01 | | Release date: | 2025-03-12 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Human TRIM21 PRYSPRY domain in complex with AL236
To Be Published
|
|
6ZIW
 
 | | The IRAK3 Pseudokinase Domain Bound To ATPgammaS | | Descriptor: | Interleukin-1 receptor-associated kinase 3, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, SULFATE ION | | Authors: | Mathea, S, Chatterjee, D, Preuss, F, Kraemer, A, Knapp, S. | | Deposit date: | 2020-06-26 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | The IRAK3 Pseudokinase Domain Bound To ATPgammaS
To Be Published
|
|
7QGP
 
 | | STK10 (LOK) bound to Macrocycle CKJB51 | | Descriptor: | 1,2-ETHANEDIOL, 1-[(4-chloranyl-2-methyl-phenyl)methyl]-3-(7,10-dioxa-13,17,18,21-tetrazatetracyclo[12.5.2.1^{2,6}.0^{17,20}]docosa-1(20),2(22),3,5,14(21),15,18-heptaen-5-yl)urea, CHLORIDE ION, ... | | Authors: | Berger, B.-T, Schroeder, M, Kraemer, A, Schwalm, M.P, Kurz, C.G, Amrhein, J.A, Hanke, T, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-12-09 | | Release date: | 2022-01-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | STK10 bound to CKJB51
To Be Published
|
|
8BK0
 
 | | Crystal structure of human Ephrin type-A receptor 2 (EPHA2) Kinase domain in complex with LDN-211904 | | Descriptor: | 1,2-ETHANEDIOL, Ephrin type-A receptor 2, ~{N}-(2-chlorophenyl)-6-piperidin-4-yl-imidazo[1,2-a]pyridine-3-carboxamide | | Authors: | Zhubi, R, Gerninghaus, J, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-08 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of human Ephrin type-A receptor 2 (EPHA2) Kinase domain in complex with LDN-211904
To Be Published
|
|
9HIN
 
 | | Crystal structure of human TRIM7 PRYSPRY domain bound to ((S)-2-(6-(6-methoxypyridin-3-yl)-1-oxoisoindolin-2-yl)-3-phenylpropanoyl)-L-glutamine | | Descriptor: | ((S)-2-(6-(6-methoxypyridin-3-yl)-1-oxoisoindolin-2-yl)-3-phenylpropanoyl)-L-glutamine, 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase TRIM7, ... | | Authors: | Lenz, C, Haman, A, Spissinger, H, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-11-26 | | Release date: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of human TRIM7 PRYSPRY domain bound to ((S)-2-(6-(6-methoxypyridin-3-yl)-1-oxoisoindolin-2-yl)-3-phenylpropanoyl)-L-glutamine
To Be Published
|
|
6T6F
 
 | | Crystal structure of human calmodulin-dependent protein kinase 1D (CAMK1D) bound to compound 8 (CS275) | | Descriptor: | 2-[(3~{S})-3-azanylpiperidin-1-yl]-4-[[3-(trifluoromethyl)phenyl]amino]pyrimidine-5-carboxamide, Calcium/calmodulin-dependent protein kinase type 1D | | Authors: | Sorrell, F, Kraemer, A, Butterworth, S, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-18 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | CAMK1D bound to CS275
To Be Published
|
|
8R5B
 
 | |
7HLP
 
 | | PanDDA analysis group deposition -- Crystal Structure of TRIM21 in complex with Z29634868 | | Descriptor: | 1,2-ETHANEDIOL, 2-oxo-4-propyl-2H-1-benzopyran-7-yl acetate, E3 ubiquitin-protein ligase TRIM21, ... | | Authors: | Kim, Y, Marples, P, Fearon, D, von Delft, F, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-11-04 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7HM5
 
 | | PanDDA analysis group deposition -- Crystal Structure of TRIM21 in complex with Z192955056 | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase TRIM21, SULFATE ION, ... | | Authors: | Kim, Y, Marples, P, Fearon, D, von Delft, F, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-11-04 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7HMK
 
 | | PanDDA analysis group deposition -- Crystal Structure of TRIM21 in complex with Z271004858 | | Descriptor: | 1,2-ETHANEDIOL, 4-amino-N-(pyridin-2-yl)benzenesulfonamide, E3 ubiquitin-protein ligase TRIM21, ... | | Authors: | Kim, Y, Marples, P, Fearon, D, von Delft, F, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-11-04 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7HN1
 
 | | PanDDA analysis group deposition -- Crystal Structure of TRIM21 in complex with Z2065616520 | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase TRIM21, N-ethyl-1-(pyridin-4-yl)piperidin-4-amine, ... | | Authors: | Kim, Y, Marples, P, Fearon, D, von Delft, F, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-11-04 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7HNH
 
 | | PanDDA analysis group deposition -- Crystal Structure of TRIM21 in complex with Z2643472210 | | Descriptor: | 1,2-ETHANEDIOL, 1-methyl-N-{[(2S)-oxolan-2-yl]methyl}-1H-pyrazole-3-carboxamide, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Kim, Y, Marples, P, Fearon, D, von Delft, F, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-11-04 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
