1F9F
 
 | | CRYSTAL STRUCTURE OF THE HPV-18 E2 DNA-BINDING DOMAIN | | Descriptor: | Regulatory protein E2, SULFATE ION | | Authors: | Kim, S.S, Tam, J, Wang, A.F, Hegde, R. | | Deposit date: | 2000-07-10 | | Release date: | 2000-11-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis of DNA target discrimination by papillomavirus E2 proteins.
J.Biol.Chem., 275, 2000
|
|
1L8R
 
 | | Structure of the Retinal Determination Protein Dachshund Reveals a DNA-Binding Motif | | Descriptor: | Dachshund | | Authors: | Kim, S.S, Zhang, R, Braunstein, S.E, Joachimiak, A, Cvekl, A, Hegde, R.S. | | Deposit date: | 2002-03-21 | | Release date: | 2002-06-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of the retinal determination protein Dachshund reveals a DNA binding motif.
Structure, 10, 2002
|
|
2OLE
 
 | | Crystal Structure Of Human Dipeptidyl Peptidase IV (DPPIV) Complex With Cyclic Hydrazine Derivatives | | Descriptor: | (2R)-4-(2-BENZOYL-1,2-DIAZEPAN-1-YL)-4-OXO-1-(2,4,5-TRIFLUOROPHENYL)BUTAN-2-AMINE, Dipeptidyl peptidase 4 | | Authors: | Kim, S.S, Ahn, J.H, Lee, J.O. | | Deposit date: | 2007-01-19 | | Release date: | 2008-01-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthesis, biological evaluation and structural determination of beta-aminoacyl-containing cyclic hydrazine derivatives as dipeptidyl peptidase IV (DPP-IV) inhibitors
Bioorg.Med.Chem.Lett., 17, 2007
|
|
4TX1
 
 | |
1BFA
 
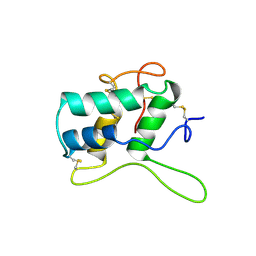 | | RECOMBINANT BIFUNCTIONAL HAGEMAN FACTOR/AMYLASE INHIBITOR FROM MAIZE | | Descriptor: | BIFUNCTIONAL AMYLASE/SERINE PROTEASE INHIBITOR | | Authors: | Behnke, C.A, Yee, V.C, Le Trong, I, Pedersen, L.C, Stenkamp, R.E, Kim, S.S, Reeck, G.R, Teller, D.C. | | Deposit date: | 1998-05-13 | | Release date: | 1998-08-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural determinants of the bifunctional corn Hageman factor inhibitor: x-ray crystal structure at 1.95 A resolution.
Biochemistry, 37, 1998
|
|
1B73
 
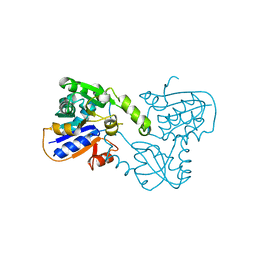 | | GLUTAMATE RACEMASE FROM AQUIFEX PYROPHILUS | | Descriptor: | GLUTAMATE RACEMASE | | Authors: | Hwang, K.Y, Cho, C.S, Kim, S.S, Yu, Y.G, Cho, Y. | | Deposit date: | 1999-01-26 | | Release date: | 1999-01-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and mechanism of glutamate racemase from Aquifex pyrophilus.
Nat.Struct.Biol., 6, 1999
|
|
1BEA
 
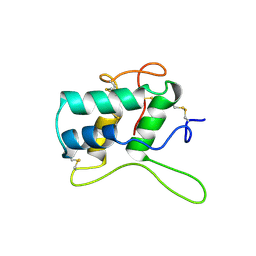 | | BIFUNCTIONAL HAGEMAN FACTOR/AMYLASE INHIBITOR FROM MAIZE | | Descriptor: | BIFUNCTIONAL AMYLASE/SERINE PROTEASE INHIBITOR | | Authors: | Behnke, C.A, Yee, V.C, Le Trong, I, Pedersen, L.C, Stenkamp, R.E, Kim, S.S, Reeck, G.R, Teller, D.C. | | Deposit date: | 1998-05-13 | | Release date: | 1998-08-12 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural determinants of the bifunctional corn Hageman factor inhibitor: x-ray crystal structure at 1.95 A resolution.
Biochemistry, 37, 1998
|
|
1B74
 
 | | GLUTAMATE RACEMASE FROM AQUIFEX PYROPHILUS | | Descriptor: | D-GLUTAMINE, GLUTAMATE RACEMASE | | Authors: | Hwang, K.Y, Cho, C.S, Kim, S.S, Yu, Y.G, Cho, Y. | | Deposit date: | 1999-01-27 | | Release date: | 2000-01-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and mechanism of glutamate racemase from Aquifex pyrophilus.
Nat.Struct.Biol., 6, 1999
|
|
3BEP
 
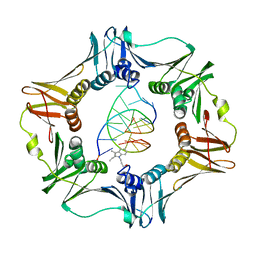 | | Structure of a sliding clamp on DNA | | Descriptor: | 1-(3-hydroxypropyl)-2-{(1E,3E,5E)-5-[1-(3-hydroxypropyl)-3,3-dimethyl-1,3-dihydro-2H-indol-2-ylidene]penta-1,3-dien-1-y l}-3,3-dimethyl-3H-indolium, DNA (5'-D(*DTP*DTP*DTP*DTP*DAP*DTP*DAP*DCP*DGP*DAP*DTP*DGP*DGP*DG)-3'), DNA (5'-D(P*DCP*DCP*DCP*DAP*DTP*DCP*DGP*DTP*DAP*DT)-3'), ... | | Authors: | Georgescu, R.E, Kim, S.S, Yurieva, O, Kuriyan, J, Kong, X.-P, O'Donnell, M. | | Deposit date: | 2007-11-19 | | Release date: | 2008-01-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure of a sliding clamp on DNA
Cell(Cambridge,Mass.), 132, 2008
|
|
1JJH
 
 | |
1JJ4
 
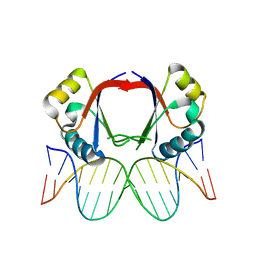 | | Human papillomavirus type 18 E2 DNA-binding domain bound to its DNA target | | Descriptor: | E2 binding site DNA, Regulatory protein E2 | | Authors: | Kim, S.-S, Tam, J.K, Wang, A.F, Hegde, R.S. | | Deposit date: | 2001-07-03 | | Release date: | 2001-07-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structural basis of DNA target discrimination by papillomavirus E2 proteins.
J.Biol.Chem., 275, 2000
|
|
1R1A
 
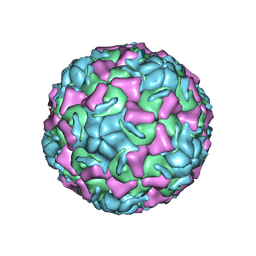 | | CRYSTAL STRUCTURE OF HUMAN RHINOVIRUS SEROTYPE 1A (HRV1A) | | Descriptor: | HUMAN RHINOVIRUS 1A COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 1A COAT PROTEIN (SUBUNIT VP2), HUMAN RHINOVIRUS 1A COAT PROTEIN (SUBUNIT VP3), ... | | Authors: | Kim, S, Rossmann, M.G. | | Deposit date: | 1989-03-15 | | Release date: | 1990-07-15 | | Last modified: | 2023-03-15 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of human rhinovirus serotype 1A (HRV1A).
J.Mol.Biol., 210, 1989
|
|
7PGB
 
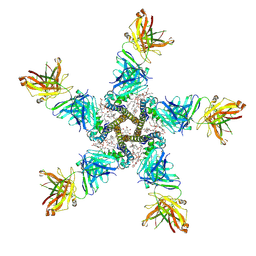 | | NaV_Ae1/Sp1CTD_pore-SAT09 complex | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 4-NITROBENZOIC ACID, ... | | Authors: | Lolicato, M, Arrigoni, C. | | Deposit date: | 2021-08-13 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Quaternary structure independent folding of voltage-gated ion channel pore domain subunits.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7PG8
 
 | | NaV_Ae1/Sp1CTD_pore-ANT05 complex | | Descriptor: | ANT05 H12 fab fragment, heavy chain, light chain, ... | | Authors: | Lolicato, M, Arrigoni, C. | | Deposit date: | 2021-08-13 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Quaternary structure independent folding of voltage-gated ion channel pore domain subunits.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7PGF
 
 | |
7PGI
 
 | | NaVAb1p (bicelles) | | Descriptor: | ACETATE ION, Ion transport protein, MAGNESIUM ION, ... | | Authors: | Lolicato, M, Arrigoni, C. | | Deposit date: | 2021-08-14 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.638 Å) | | Cite: | Quaternary structure independent folding of voltage-gated ion channel pore domain subunits.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7PGH
 
 | | NaVAe1/Sp1CTDp (DDM) | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, DODECAETHYLENE GLYCOL, DODECANE, ... | | Authors: | Lolicato, M, Arrigoni, C. | | Deposit date: | 2021-08-14 | | Release date: | 2022-06-15 | | Last modified: | 2022-06-29 | | Method: | X-RAY DIFFRACTION (4.194 Å) | | Cite: | Quaternary structure independent folding of voltage-gated ion channel pore domain subunits.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7PGG
 
 | | NaVAb1p detergent (DM) | | Descriptor: | 2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}ethyl heptadecanoate, Ion transport protein | | Authors: | Lolicato, M, Arrigoni, C. | | Deposit date: | 2021-08-13 | | Release date: | 2022-06-15 | | Last modified: | 2022-06-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Quaternary structure independent folding of voltage-gated ion channel pore domain subunits.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7CM4
 
 | | Crystal Structure of COVID-19 virus spike receptor-binding domain complexed with a neutralizing antibody CT-P59 | | Descriptor: | 1,2-ETHANEDIOL, IgG heavy chain, IgG light chain, ... | | Authors: | Kim, Y.G, Jeong, J.H, Bae, J.S, Lee, J. | | Deposit date: | 2020-07-24 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | A therapeutic neutralizing antibody targeting receptor binding domain of SARS-CoV-2 spike protein.
Nat Commun, 12, 2021
|
|
5BK1
 
 | |
5BK2
 
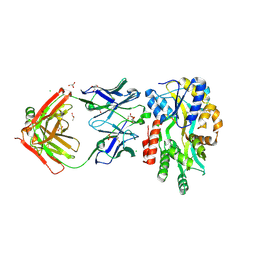 | |
5BJZ
 
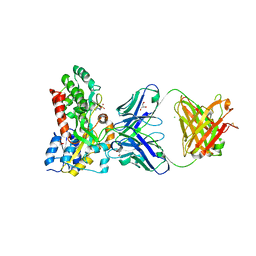 | |
3EIO
 
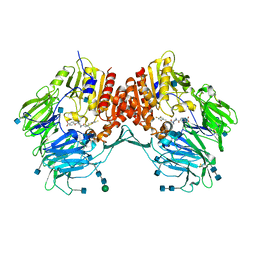 | | Crystal Structure Analysis of DPPIV Inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-({4-[(3R)-3-amino-4-(2,4,5-trifluorophenyl)butanoyl]-1,4-diazepan-1-yl}carbonyl)benzoic acid, ... | | Authors: | Ahn, J.H, Lee, J.-O. | | Deposit date: | 2008-09-17 | | Release date: | 2008-11-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis and biological evaluation of homopiperazine derivatives with beta-aminoacyl group as dipeptidyl peptidase IV inhibitors
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3Q8W
 
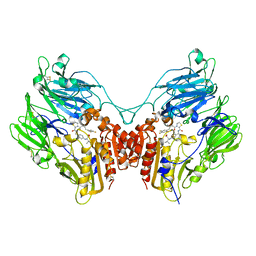 | | A b-aminoacyl containing thiazolidine derivative and DPPIV complex | | Descriptor: | Dipeptidyl peptidase 4, N-(4-{[({(2R)-3-[(3R)-3-amino-4-(2,4,5-trifluorophenyl)butanoyl]-1,3-thiazolidin-2-yl}carbonyl)amino]methyl}phenyl)-D-valine | | Authors: | Lee, J.O, Song, D.H. | | Deposit date: | 2011-01-07 | | Release date: | 2011-03-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.64 Å) | | Cite: | Discovery of b-aminoacyl containing thiazolidine derivatives as potent and selective dipeptidyl peptidase IV inhibitors
To be Published
|
|
7COE
 
 | | Crystal structure of Receptor binding domain of MERS-CoV and KNIH90-F1 Fab complex | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain, ... | | Authors: | Lee, J.Y, Song, J.Y, Lee, H.S, Hong, E, Jang, T.H. | | Deposit date: | 2020-08-04 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The structure of a novel antibody against the spike protein inhibits Middle East respiratory syndrome coronavirus infections.
Sci Rep, 12, 2022
|
|
