5VC7
 
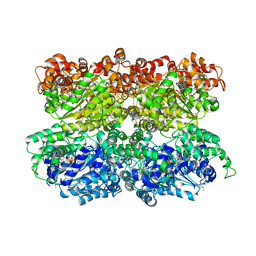 | | VCP like ATPase from T. acidophilum (VAT) - conformation 1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, VCP-like ATPase | | Authors: | Ripstein, Z.A, Huang, R, Augustyniak, R, Kay, L.E, Rubinstein, J.L. | | Deposit date: | 2017-03-31 | | Release date: | 2017-04-26 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of a AAA+ unfoldase in the process of unfolding substrate.
Elife, 6, 2017
|
|
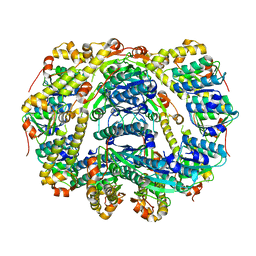
6DKF
 
 | |
5VCA
 
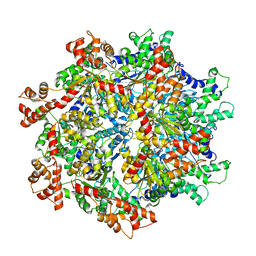 | | VCP like ATPase from T. acidophilum (VAT)-Substrate bound conformation | | Descriptor: | VCP-like ATPase | | Authors: | Ripstein, Z.A, Huang, R, Augustyniak, R, Kay, L.E, Rubinstein, J.L. | | Deposit date: | 2017-03-31 | | Release date: | 2017-04-26 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure of a AAA+ unfoldase in the process of unfolding substrate.
Elife, 6, 2017
|
|
8UX1
 
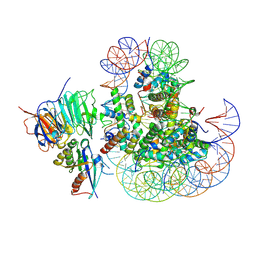 | |
1Y8B
 
 | |
1DDM
 
 | | SOLUTION STRUCTURE OF THE NUMB PTB DOMAIN COMPLEXED TO A NAK PEPTIDE | | Descriptor: | NUMB ASSOCIATE KINASE, NUMB PROTEIN | | Authors: | Zwahlen, C, Li, S.C, Kay, L.E, Pawson, T, Forman-Kay, J.D. | | Deposit date: | 1999-11-11 | | Release date: | 2000-04-10 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Multiple modes of peptide recognition by the PTB domain of the cell fate determinant Numb.
EMBO J., 19, 2000
|
|
5G4F
 
 | | Structure of the ADP-bound VAT complex | | Descriptor: | VCP-LIKE ATPASE | | Authors: | Huang, R, Ripstein, Z.A, Augustyniak, R, Lazniewski, M, Ginalski, K, Kay, L.E, Rubinstein, J.L. | | Deposit date: | 2016-05-12 | | Release date: | 2016-07-27 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Unfolding the Mechanism of the Aaa+ Unfoldase Vat by a Combined Cryo-Em, Solution NMR Study.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5G4G
 
 | | Structure of the ATPgS-bound VAT complex | | Descriptor: | VCP-LIKE ATPASE | | Authors: | Huang, R, Ripstein, Z.A, Augustyniak, R, Lazniewski, M, Ginalski, K, Kay, L.E, Rubinstein, J.L. | | Deposit date: | 2016-05-12 | | Release date: | 2016-07-27 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Unfolding the Mechanism of the Aaa+ Unfoldase Vat by a Combined Cryo-Em, Solution NMR Study.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2JQX
 
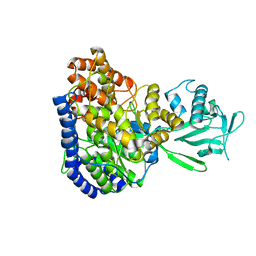 | | Solution structure of Malate Synthase G from joint refinement against NMR and SAXS data | | Descriptor: | Malate synthase G | | Authors: | Grishaev, A, Tugarinov, V, Kay, L.E, Trewhella, J, Bax, A. | | Deposit date: | 2007-06-13 | | Release date: | 2007-07-10 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of the 82-kDa enzyme malate synthase G from joint NMR and synchrotron SAXS restraints
J.Biomol.Nmr, 40, 2008
|
|
2IL6
 
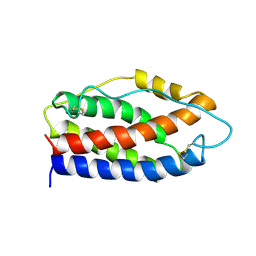 | | HUMAN INTERLEUKIN-6, NMR, 32 STRUCTURES | | Descriptor: | INTERLEUKIN-6 | | Authors: | Xu, G.Y, Yu, H.A, Hong, J, Stahl, M, Mcdonagh, T, Kay, L.E, Cumming, D.A. | | Deposit date: | 1997-01-31 | | Release date: | 1998-02-04 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of recombinant human interleukin-6.
J.Mol.Biol., 268, 1997
|
|
1KA7
 
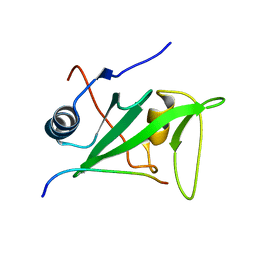 | | SAP/SH2D1A bound to peptide n-Y-c | | Descriptor: | SH2 DOMAIN PROTEIN 1A, peptide n-Y-c | | Authors: | Hwang, P.M, Li, C, Morra, M, Lillywhite, J, Gertler, F, Terhorst, C, Kay, L.E, Pawson, T, Forman-Kay, J, Li, S.-C. | | Deposit date: | 2001-10-31 | | Release date: | 2001-11-07 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | A "three-pronged" binding mechanism for the SAP/SH2D1A SH2 domain: structural basis and relevance to the XLP syndrome.
EMBO J., 21, 2002
|
|
1KA6
 
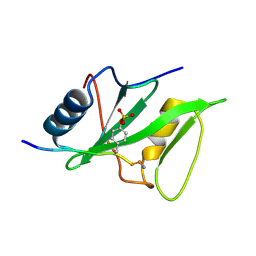 | | SAP/SH2D1A bound to peptide n-pY | | Descriptor: | SH2 DOMAIN PROTEIN 1A, peptide n-pY | | Authors: | Hwang, P.M, Li, C, Morra, M, Lillywhite, J, Gertler, F, Terhorst, C, Kay, L.E, Pawson, T, Forman-Kay, J, Li, S.-C. | | Deposit date: | 2001-10-31 | | Release date: | 2001-11-07 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | A "three-pronged" binding mechanism for the SAP/SH2D1A SH2 domain: structural basis and relevance to the XLP syndrome.
EMBO J., 21, 2002
|
|
1MM4
 
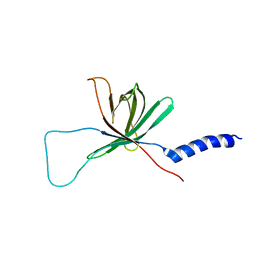 | | Solution NMR structure of the outer membrane enzyme PagP in DPC micelles | | Descriptor: | CrcA protein | | Authors: | Hwang, P.M, Choy, W.-Y, Lo, E.I, Chen, L, Forman-Kay, J.D, Raetz, C.R.H, Prive, G.G, Bishop, R.E, Kay, L.E. | | Deposit date: | 2002-09-03 | | Release date: | 2002-09-13 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Dynamics of the Outer Membrane Enzyme PagP by NMR
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1EZO
 
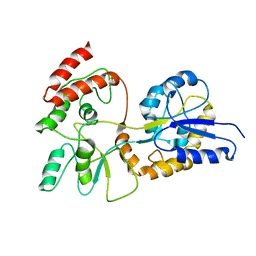 | | GLOBAL FOLD OF MALTODEXTRIN BINDING PROTEIN COMPLEXED WITH BETA-CYCLODEXTRIN | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN | | Authors: | Mueller, G.A, Choy, W.Y, Yang, D, Forman-Kay, J.D, Venters, R.A, Kay, L.E. | | Deposit date: | 2000-05-11 | | Release date: | 2001-05-03 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Global folds of proteins with low densities of NOEs using residual dipolar couplings: application to the 370-residue maltodextrin-binding protein.
J.Mol.Biol., 300, 2000
|
|
1EZP
 
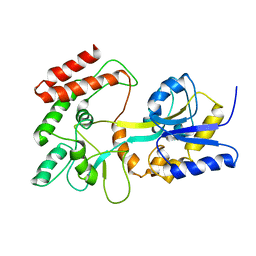 | | GLOBAL FOLD OF MALTODEXTRIN BINDING PROTEIN COMPLEXED WITH BETA-CYCLODEXTRIN USING PEPTIDE ORIENTATIONS FROM DIPOLAR COUPLINGS | | Descriptor: | MALTODEXTRIN BINDING PERIPLASMIC PROTEIN | | Authors: | Mueller, G.A, Choy, W.Y, Yang, D, Forman-Kay, J.D, Venters, R.A, Kay, L.E. | | Deposit date: | 2000-05-11 | | Release date: | 2001-05-08 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Global folds of proteins with low densities of NOEs using residual dipolar couplings: application to the 370-residue maltodextrin-binding protein.
J.Mol.Biol., 300, 2000
|
|
1MM5
 
 | | Solution NMR structure of the outer membrane enzyme PagP in OG micelles | | Descriptor: | CrcA protein | | Authors: | Hwang, P.M, Choy, W.-Y, Lo, E.I, Chen, L, Forman-Kay, J.D, Raetz, C.R.H, Prive, G.G, Bishop, R.E, Kay, L.E. | | Deposit date: | 2002-09-03 | | Release date: | 2002-09-13 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Dynamics of the Outer Membrane Enzyme PagP by NMR
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1JU5
 
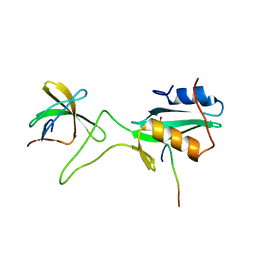 | | Ternary complex of an Crk SH2 domain, Crk-derived phophopeptide, and Abl SH3 domain by NMR spectroscopy | | Descriptor: | Abl, Crk | | Authors: | Donaldson, L.W, Pawson, T, Kay, L.E, Forman-Kay, J.D. | | Deposit date: | 2001-08-23 | | Release date: | 2002-11-06 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Structure of a regulatory complex involving the Abl SH3 domain, the Crk
SH2 domain, and a Crk-derived phosphopeptide
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2KIS
 
 | | Solution structure of CA150 FF1 domain and FF1-FF2 interdomain linker | | Descriptor: | Transcription elongation regulator 1 | | Authors: | Murphy, J.M, Hansen, D, Wiesner, S, Muhandiram, D, Borg, M, Smith, M.J, Sicheri, F, Kay, L.E, Forman-Kay, J.D, Pawson, T. | | Deposit date: | 2009-05-08 | | Release date: | 2009-09-08 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural studies of FF domains of the transcription factor CA150 provide insights into the organization of FF domain tandem arrays.
J.Mol.Biol., 393, 2009
|
|
2PLE
 
 | | NUCLEAR MAGNETIC RESONANCE STRUCTURE OF AN SH2 DOMAIN OF PHOSPHOLIPASE C-GAMMA1 COMPLEXED WITH A HIGH AFFINITY BINDING PEPTIDE | | Descriptor: | PHOSPHOLIPASE C GAMMA-1, C-TERMINAL SH2 DOMAIN, PHOSPHOPEPTIDE FROM PDGF | | Authors: | Pascal, S.M, Singer, A.U, Gish, G, Yamazaki, T, Shoelson, S.E, Pawson, T, Kay, L.E, Forman-Kay, J.D. | | Deposit date: | 1994-08-19 | | Release date: | 1995-01-26 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure of an SH2 domain of phospholipase C-gamma 1 complexed with a high affinity binding peptide.
Cell(Cambridge,Mass.), 77, 1994
|
|
2PLD
 
 | | NUCLEAR MAGNETIC RESONANCE STRUCTURE OF AN SH2 DOMAIN OF PHOSPHOLIPASE C-GAMMA1 COMPLEXED WITH A HIGH AFFINITY BINDING PEPTIDE | | Descriptor: | PHOSPHOLIPASE C GAMMA-1, C-TERMINAL SH2 DOMAIN, PHOSPHOPEPTIDE FROM PDGF | | Authors: | Pascal, S.M, Singer, A.U, Gish, G, Yamazaki, T, Shoelson, S.E, Pawson, T, Kay, L.E, Forman-Kay, J.D. | | Deposit date: | 1994-08-19 | | Release date: | 1995-01-26 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure of an SH2 domain of phospholipase C-gamma 1 complexed with a high affinity binding peptide.
Cell(Cambridge,Mass.), 77, 1994
|
|
1IL6
 
 | | HUMAN INTERLEUKIN-6, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | INTERLEUKIN-6 | | Authors: | Xu, G.Y, Yu, H.A, Hong, J, Stahl, M, Mcdonagh, T, Kay, L.E, Cumming, D.A. | | Deposit date: | 1997-01-31 | | Release date: | 1998-02-04 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of recombinant human interleukin-6.
J.Mol.Biol., 268, 1997
|
|
8F0U
 
 | | Structure of a 12mer DegP cage bound to the client protein hTRF1 | | Descriptor: | Periplasmic serine endoprotease DegP, Telomeric repeat-binding factor 1 | | Authors: | Harkness, R.W, Ripstein, Z.A, Di Trani, J.M, Kay, L.E. | | Deposit date: | 2022-11-04 | | Release date: | 2022-11-23 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Flexible Client-Dependent Cages in the Assembly Landscape of the Periplasmic Protease-Chaperone DegP.
J.Am.Chem.Soc., 145, 2023
|
|
8F26
 
 | | Structure of a 60mer DegP cage bound to the client protein hTRF1 | | Descriptor: | Periplasmic serine endoprotease DegP, Telomeric repeat-binding factor 1 | | Authors: | Harkness, R.W, Ripstein, Z.A, Di Trani, J.M, Kay, L.E. | | Deposit date: | 2022-11-07 | | Release date: | 2022-11-23 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (9.7 Å) | | Cite: | Flexible Client-Dependent Cages in the Assembly Landscape of the Periplasmic Protease-Chaperone DegP.
J.Am.Chem.Soc., 145, 2023
|
|
8F21
 
 | | Structure of a 30mer DegP cage bound to the client protein hTRF1 | | Descriptor: | Periplasmic serine endoprotease DegP, Telomeric repeat-binding factor 1 | | Authors: | Harkness, R.W, Ripstein, Z.A, Di Trani, J.M, Kay, L.E. | | Deposit date: | 2022-11-06 | | Release date: | 2022-11-23 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (14.1 Å) | | Cite: | Flexible Client-Dependent Cages in the Assembly Landscape of the Periplasmic Protease-Chaperone DegP.
J.Am.Chem.Soc., 145, 2023
|
|
8F0A
 
 | | Client-bound structure of a DegP trimer within a 12mer cage | | Descriptor: | Periplasmic serine endoprotease DegP, Telomeric repeat-binding factor 1 | | Authors: | Harkness, R.W, Ripstein, Z.A, Di Trani, J.M, Kay, L.E. | | Deposit date: | 2022-11-02 | | Release date: | 2022-11-23 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Flexible Client-Dependent Cages in the Assembly Landscape of the Periplasmic Protease-Chaperone DegP.
J.Am.Chem.Soc., 145, 2023
|
|
