1ATW
 
 | | HAIRPIN WITH AGAU TETRALOOP, NMR, 3 STRUCTURES | | Descriptor: | RNA (5'-R(*GP*CP*UP*CP*CP*AP*GP*AP*UP*GP*GP*AP*GP*CP*G)-3') | | Authors: | Kang, H. | | Deposit date: | 1997-08-14 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Primary Sequence at the Junction of Stem and Loop in RNA Hairpins Affects the Three-Dimensional Conformation in Solution
To be Published
|
|
1ATV
 
 | | HAIRPIN WITH AGAA TETRALOOP, NMR, 4 STRUCTURES | | Descriptor: | RNA (5'-R(*GP*GP*GP*AP*CP*CP*AP*GP*AP*AP*GP*GP*UP*CP*CP*CP*G)-3') | | Authors: | Kang, H. | | Deposit date: | 1997-08-14 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Primary Sequence at the Junction of Stem and Loop in RNA Hairpins Affects the Three-Dimensional Conformation in Solution
To be Published
|
|
1KPD
 
 | |
7YON
 
 | | Complex structure of Neuropeptide Y Y2 receptor in complex with PYY(3-36) and Gi | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Kang, H, Park, C, Kim, J, Choi, H.-J. | | Deposit date: | 2022-08-01 | | Release date: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis for Y2 receptor-mediated neuropeptide Y and peptide YY signaling.
Structure, 31, 2023
|
|
7YOO
 
 | | Complex structure of Neuropeptide Y Y2 receptor in complex with NPY and Gi | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Kang, H, Park, C, Kim, J, Choi, H.-J. | | Deposit date: | 2022-08-01 | | Release date: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural basis for Y2 receptor-mediated neuropeptide Y and peptide YY signaling.
Structure, 31, 2023
|
|
1KAJ
 
 | |
5H5M
 
 | | Crystal structure of HMP-1 M domain | | Descriptor: | Alpha-catenin-like protein hmp-1 | | Authors: | Kang, H, Bang, I, Weis, W.I, Choi, H.J. | | Deposit date: | 2016-11-08 | | Release date: | 2017-03-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional characterization of Caenorhabditis elegans alpha-catenin reveals constitutive binding to beta-catenin and F-actin
J. Biol. Chem., 292, 2017
|
|
6KL7
 
 | | Beta-arrestin 1 mutant S13D/T275D | | Descriptor: | 1,2-ETHANEDIOL, BARIUM ION, Beta-arrestin-1 | | Authors: | Kang, H, Choi, H.J. | | Deposit date: | 2019-07-29 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Conformational Dynamics and Functional Implications of Phosphorylated beta-Arrestins.
Structure, 28, 2020
|
|
8E96
 
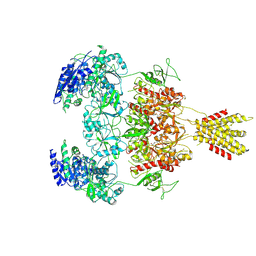 | | Glycine and glutamate bound Human GluN1a-GluN2D NMDA receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, GLYCINE, ... | | Authors: | Kang, H, Furukawa, H. | | Deposit date: | 2022-08-26 | | Release date: | 2022-12-07 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural insights into assembly and function of GluN1-2C, GluN1-2A-2C, and GluN1-2D NMDARs.
Mol.Cell, 82, 2022
|
|
5J2Y
 
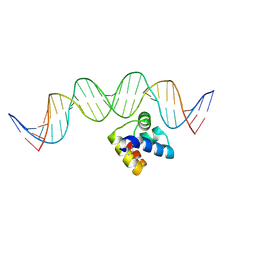 | | Molecular insight into the regulatory mechanism of the quorum-sensing repressor RsaL in Pseudomonas aeruginosa | | Descriptor: | DNA (26-MER), Regulatory protein | | Authors: | Zhao, J, Gan, J, Zhang, J, Kang, H, Kong, W, Zhu, M, Li, F, Song, Y, Qin, J, Liang, H. | | Deposit date: | 2016-03-30 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Pseudomonas aeruginosa RsaL bound to promoter DNA reaffirms its role as a global regulator involved in quorum-sensing.
Nucleic Acids Res., 45, 2017
|
|
2JV3
 
 | |
8HTG
 
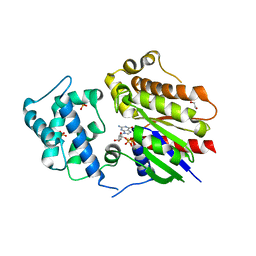 | | Crystal structure of Golf in complex with GTP-gamma S and Mg | | Descriptor: | 1,2-ETHANEDIOL, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Guanine nucleotide-binding protein G(olf) subunit alpha, ... | | Authors: | Kang, H, Choi, H.-J. | | Deposit date: | 2022-12-21 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Understanding the molecular mechanisms of odorant binding and activation of the human OR52 family.
Nat Commun, 14, 2023
|
|
5XA5
 
 | | Crystal structure of HMP-1-HMP-2 complex | | Descriptor: | Alpha-catenin-like protein hmp-1, Beta-catenin-like protein hmp-2 | | Authors: | Shao, X, Kang, H, Weis, W.I, Hardin, J, Choi, H.J. | | Deposit date: | 2017-03-11 | | Release date: | 2017-08-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cell-cell adhesion in metazoans relies on evolutionarily conserved features of the alpha-catenin· beta-catenin-binding interface.
J.Biol.Chem., 292, 2017
|
|
7VGX
 
 | | Neuropeptide Y Y1 Receptor (NPY1R) in Complex with G Protein and its endogeneous Peptide-Agonist Neuropeptide Y (NPY) | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Park, C, Kim, J, Jeong, H, Kang, H, Bang, I, Choi, H.-J. | | Deposit date: | 2021-09-19 | | Release date: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of neuropeptide Y signaling through Y1 receptor
Nat Commun, 13, 2022
|
|
7XSG
 
 | | Crystal structure of ClAgl29B | | Descriptor: | Alpha-L-fucosidase, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shishiuchi, R, Kang, H, Tagami, T, Okuyama, M. | | Deposit date: | 2022-05-14 | | Release date: | 2023-01-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.609 Å) | | Cite: | Discovery of alpha-l-Glucosidase Raises the Possibility of alpha-l-Glucosides in Nature.
Acs Omega, 7, 2022
|
|
7XSF
 
 | | Crystal structure of ClAgl29A | | Descriptor: | Alpha-L-fucosidase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Shishiuchi, R, Kang, H, Tagami, T, Okuyama, M. | | Deposit date: | 2022-05-14 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Discovery of alpha-l-Glucosidase Raises the Possibility of alpha-l-Glucosides in Nature.
Acs Omega, 7, 2022
|
|
7XSH
 
 | | Crystal structure of ClAgl29B bound with L-glucose | | Descriptor: | Alpha-L-fucosidase, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shishiuchi, R, Kang, H, Tagami, T, Okuyama, M. | | Deposit date: | 2022-05-14 | | Release date: | 2023-01-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.708 Å) | | Cite: | Discovery of alpha-l-Glucosidase Raises the Possibility of alpha-l-Glucosides in Nature.
Acs Omega, 7, 2022
|
|
4ZZ7
 
 | | Crystal structure of methylmalonate-semialdehyde dehydrogenase (DddC) from Oceanimonas doudoroffii | | Descriptor: | Methylmalonate-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Do, H, Lee, C.W, Lee, S.G, Kang, H, Park, C.M, Kim, H.J, Park, H, Park, H, Lee, J.H. | | Deposit date: | 2015-05-22 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure and modeling of the tetrahedral intermediate state of methylmalonate-semialdehyde dehydrogenase (MMSDH) from Oceanimonas doudoroffii.
J. Microbiol., 54, 2016
|
|
8KB4
 
 | | Cryo-EM structure of human TMEM87A A308M | | Descriptor: | (9R,12R)-15-amino-12-hydroxy-6,12-dioxo-7,11,13-trioxa-12lambda~5~-phosphapentadecan-9-yl undecanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Transmembrane protein 87A,EGFP | | Authors: | Han, A, Kim, H.M. | | Deposit date: | 2023-08-03 | | Release date: | 2024-07-10 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | GolpHCat (TMEM87A), a unique voltage-dependent cation channel in Golgi apparatus, contributes to Golgi-pH maintenance and hippocampus-dependent memory.
Nat Commun, 15, 2024
|
|
2JUO
 
 | | GABPa OST domain | | Descriptor: | GA-binding protein alpha chain | | Authors: | Kang, H, Nelson, M.L, Mackereth, C.D, Schaerpf, M, Graves, B.J, McIntosh, L.P. | | Deposit date: | 2007-08-31 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Identification and structural characterization of a CBP/p300-binding domain from the ETS family transcription factor GABP alpha
J.Mol.Biol., 377, 2008
|
|
8HSI
 
 | | Cryo-EM structure of human TMEM87A, PE-bound | | Descriptor: | (1S)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Han, A, Kim, H.M. | | Deposit date: | 2022-12-19 | | Release date: | 2023-12-27 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | GolpHCat (TMEM87A), a unique voltage-dependent cation channel in Golgi apparatus, contributes to Golgi-pH maintenance and hippocampus-dependent memory.
Nat Commun, 15, 2024
|
|
8HTT
 
 | | Cryo-EM structure of human TMEM87A, gluconate-bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, D-gluconic acid, Transmembrane protein 87A,EGFP, ... | | Authors: | Han, A, Kim, H.M. | | Deposit date: | 2022-12-21 | | Release date: | 2023-12-27 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | GolpHCat (TMEM87A), a unique voltage-dependent cation channel in Golgi apparatus, contributes to Golgi-pH maintenance and hippocampus-dependent memory.
Nat Commun, 15, 2024
|
|
5DZZ
 
 | |
7EJU
 
 | | Junin virus(JUNV) RNA polymerase L complexed with Z protein | | Descriptor: | MAGNESIUM ION, RING finger protein Z, RNA-directed RNA polymerase L, ... | | Authors: | Chen, Y. | | Deposit date: | 2021-04-02 | | Release date: | 2021-07-07 | | Last modified: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for recognition and regulation of arenavirus polymerase L by Z protein.
Nat Commun, 12, 2021
|
|
2KMD
 
 | | Ras signaling requires dynamic properties of Ets1 for phosphorylation-enhanced binding to co-activator CBP | | Descriptor: | Protein C-ets-1 | | Authors: | Nelson, M.L, Kang, H, Lee, G.M, Blaszczak, A.G, Lau, D.K.W, McIntosh, L.P, Graves, B.J. | | Deposit date: | 2009-07-27 | | Release date: | 2010-05-05 | | Last modified: | 2012-03-21 | | Method: | SOLUTION NMR | | Cite: | Ras signaling requires dynamic properties of Ets1 for phosphorylation-enhanced binding to coactivator CBP.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
