7RKC
 
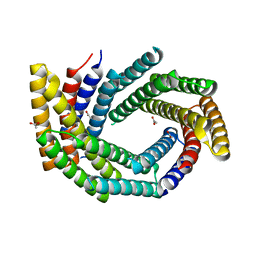 | | Computationally designed tunable C2 symmetric tandem repeat homodimer, D_3_633 | | Descriptor: | ACETATE ION, D_3_633 | | Authors: | Kennedy, M.A, Stoddard, B.L, Hicks, D.R, Bera, A.K. | | Deposit date: | 2021-07-22 | | Release date: | 2022-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | De novo design of protein homodimers containing tunable symmetric protein pockets.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RMY
 
 | | De Novo designed tunable protein pockets, D_3-337 | | Descriptor: | De Novo designed tunable homodimer, D_3-337 | | Authors: | Bera, A.K, Hicks, D.R, Kang, A, Sankaran, B, Baker, D. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | De novo design of protein homodimers containing tunable symmetric protein pockets.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RMX
 
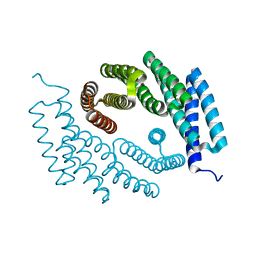 | | Structure of De Novo designed tunable symmetric protein pockets | | Descriptor: | Tunable symmetric protein, D_3_212 | | Authors: | Bera, A.K, Hicks, D.R, Kang, A, Sankaran, B, Baker, D. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | De novo design of protein homodimers containing tunable symmetric protein pockets.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5U05
 
 | |
8F6R
 
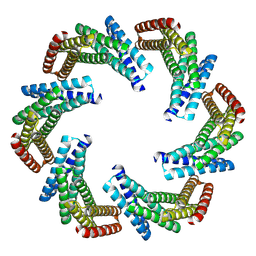 | | CryoEM structure of designed modular protein oligomer C6-79 | | Descriptor: | De novo designed oligomeric protein C6-79 | | Authors: | Redler, R.L, Edman, N.I, Baker, D, Ekiert, D, Bhabha, G. | | Deposit date: | 2022-11-17 | | Release date: | 2023-11-29 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Modulation of FGF pathway signaling and vascular differentiation using designed oligomeric assemblies.
Cell, 187, 2024
|
|
8F6Q
 
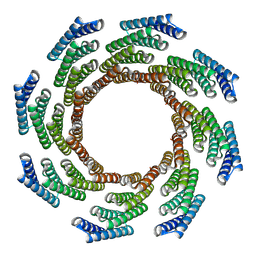 | | CryoEM structure of designed modular protein oligomer C8-71 | | Descriptor: | C8-71 | | Authors: | Redler, R.L, Edman, N.I, Baker, D, Ekiert, D, Bhabha, G. | | Deposit date: | 2022-11-17 | | Release date: | 2023-11-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Modulation of FGF pathway signaling and vascular differentiation using designed oligomeric assemblies.
Biorxiv, 2023
|
|
8DT0
 
 | |
8E55
 
 | |
8GLT
 
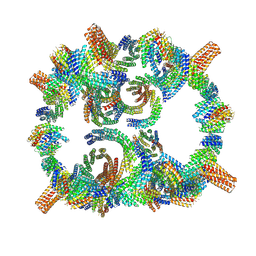 | | Backbone model of de novo-designed chlorophyll-binding nanocage O32-15 | | Descriptor: | C2-chlorophyll-comp_O32-15_ctermHis, polyalanine model, C3-comp_O32-15 | | Authors: | Redler, R.L, Ennist, N.M, Wang, S, Baker, D, Ekiert, D.C, Bhabha, G. | | Deposit date: | 2023-03-23 | | Release date: | 2024-03-27 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | De novo design of proteins housing excitonically coupled chlorophyll special pairs.
Nat.Chem.Biol., 20, 2024
|
|
8G4K
 
 | |
8GAB
 
 | |
8GAC
 
 | |
8GAD
 
 | |
8VEI
 
 | |
8VEJ
 
 | | De novo designed cholic acid binder: CHD_buttress | | Descriptor: | CHD_buttress, CHOLIC ACID | | Authors: | Bera, A.K, Tran, L, Kang, A, Baker, D. | | Deposit date: | 2023-12-19 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Binding and sensing diverse small molecules using shape-complementary pseudocycles.
Science, 385, 2024
|
|
8E1E
 
 | |
8EVM
 
 | |
8FJE
 
 | | The five-repeat design E8 | | Descriptor: | E8 | | Authors: | Bera, A.K, An, L, Baker, D. | | Deposit date: | 2022-12-19 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Hallucination of closed repeat proteins containing central pockets.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8FJF
 
 | | The three-repeat design H10 | | Descriptor: | H10 | | Authors: | Bera, A.K, An, L, Baker, D. | | Deposit date: | 2022-12-19 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Hallucination of closed repeat proteins containing central pockets.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8FJG
 
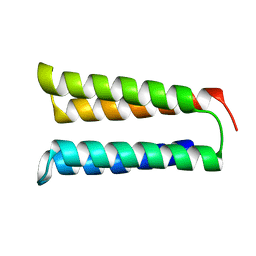 | | The two-repeat design H12 | | Descriptor: | H12 | | Authors: | Bera, A.K, An, L, Baker, D. | | Deposit date: | 2022-12-19 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Hallucination of closed repeat proteins containing central pockets.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8FRE
 
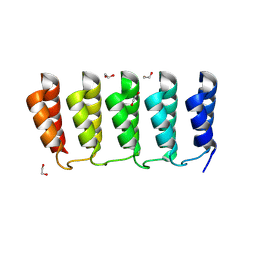 | | Designed loop protein RBL4 | | Descriptor: | 1,2-ETHANEDIOL, RBL4 | | Authors: | Jude, K.M, Jiang, H, Baker, D, Garcia, K.C. | | Deposit date: | 2023-01-06 | | Release date: | 2024-04-17 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | De novo design of buttressed loops for sculpting protein functions.
Nat.Chem.Biol., 20, 2024
|
|
8FRF
 
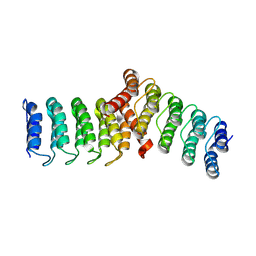 | | Homodimeric designed loop protein RBL7_C2_3 | | Descriptor: | MALONATE ION, RBL7_C2_3 | | Authors: | Jude, K.M, Jiang, H, Baker, D, Garcia, K.C. | | Deposit date: | 2023-01-06 | | Release date: | 2024-04-17 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | De novo design of buttressed loops for sculpting protein functions.
Nat.Chem.Biol., 20, 2024
|
|
5TKV
 
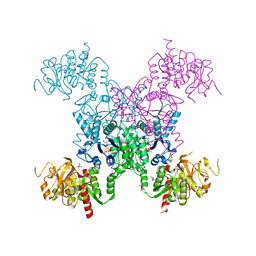 | | X-RAY CRYSTAL STRUCTURE OF THE "CLOSED" CONFORMATION OF CTP-INHIBITED E. COLI CYTIDINE TRIPHOSPHATE (CTP) SYNTHETASE | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CTP synthase, ... | | Authors: | Baldwin, E.P, Endrizzi, J.A. | | Deposit date: | 2016-10-07 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Human CTP synthase filament structure reveals the active enzyme conformation.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5U03
 
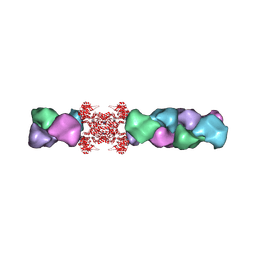 | | Cryo-EM structure of the human CTP synthase filament | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CTP synthase 1, URIDINE 5'-TRIPHOSPHATE | | Authors: | Lynch, E.M, Kollman, J.M. | | Deposit date: | 2016-11-22 | | Release date: | 2017-04-26 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Human CTP synthase filament structure reveals the active enzyme conformation.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5U3C
 
 | |
