1NAR
 
 | |
1A53
 
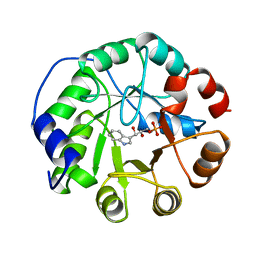 | | COMPLEX OF INDOLE-3-GLYCEROLPHOSPHATE SYNTHASE FROM SULFOLOBUS SOLFATARICUS WITH INDOLE-3-GLYCEROLPHOSPHATE AT 2.0 A RESOLUTION | | Descriptor: | INDOLE-3-GLYCEROL PHOSPHATE, INDOLE-3-GLYCEROLPHOSPHATE SYNTHASE | | Authors: | Hennig, M, Darimont, B, Kirschner, K, Jansonius, J.N. | | Deposit date: | 1998-02-19 | | Release date: | 1999-03-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The catalytic mechanism of indole-3-glycerol phosphate synthase: crystal structures of complexes of the enzyme from Sulfolobus solfataricus with substrate analogue, substrate, and product.
J.Mol.Biol., 319, 2002
|
|
4GSA
 
 | |
3GSB
 
 | |
2GSA
 
 | |
1NSJ
 
 | |
1CNV
 
 | | CRYSTAL STRUCTURE OF CONCANAVALIN B AT 1.65 A RESOLUTION | | Descriptor: | CONCANAVALIN B | | Authors: | Hennig, M. | | Deposit date: | 1995-02-20 | | Release date: | 1996-03-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of concanavalin B at 1.65 A resolution. An "inactivated" chitinase from seeds of Canavalia ensiformis.
J.Mol.Biol., 254, 1995
|
|
1LBL
 
 | | Crystal structure of indole-3-glycerol phosphate synthase (IGPS) in complex with 1-(o-carboxyphenylamino)-1-deoxyribulose 5'-phosphate (CdRP) | | Descriptor: | 1-(O-CARBOXY-PHENYLAMINO)-1-DEOXY-D-RIBULOSE-5-PHOSPHATE, indole-3-glycerol phosphate synthase | | Authors: | Hennig, M, Darimont, B.D, Kirschner, K, Jansonius, J.N. | | Deposit date: | 2002-04-04 | | Release date: | 2002-09-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The catalytic mechanism of indole-3-glycerol phosphate synthase: Crystal structures of complexes of the enzyme from Sulfolobus solfataricus with the substrate analogue, substrate, and product
J.Mol.Biol., 319, 2002
|
|
1LBF
 
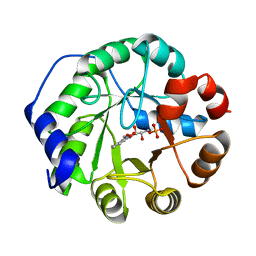 | | CRYSTAL STRUCTURE OF INDOLE-3-GLYCEROL PHOSPHATE SYNTASE (IGPS)WITH REDUCED 1-(O-CABOXYPHENYLAMINO)-1-DEOXYRIBULOSE 5-PHOSPHATE (RCDRP) | | Descriptor: | 1-(O-CARBOXY-PHENYLAMINO)-1-DEOXY-D-RIBULOSE-5-PHOSPHATE, INDOLE-3-GLYCEROL PHOSPHATE SYNTHASE | | Authors: | Hennig, M, Darimont, B, Kirschner, K, Jansonius, J.N. | | Deposit date: | 2002-04-03 | | Release date: | 2002-06-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The catalytic mechanism of indole-3-glycerol phosphate synthase: crystal structures of complexes of the enzyme from Sulfolobus solfataricus with substrate analogue, substrate, and product.
J.Mol.Biol., 319, 2002
|
|
2DHN
 
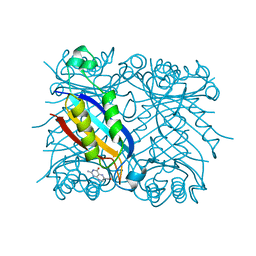 | | COMPLEX OF 7,8-DIHYDRONEOPTERIN ALDOLASE FROM STAPHYLOCOCCUS AUREUS WITH 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN AT 2.2 A RESOLUTION | | Descriptor: | 2-AMINO-6-HYDROXYMETHYL-7,8-DIHYDRO-3H-PTERIDIN-4-ONE, 7,8-DIHYDRONEOPTERIN ALDOLASE | | Authors: | Hennig, M, D'Arcy, A, Hampele, I.C, Page, M.G.P, Oefner, C.H, Dale, G. | | Deposit date: | 1998-03-31 | | Release date: | 1999-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and reaction mechanism of 7,8-dihydroneopterin aldolase from Staphylococcus aureus.
Nat.Struct.Biol., 5, 1998
|
|
1DHN
 
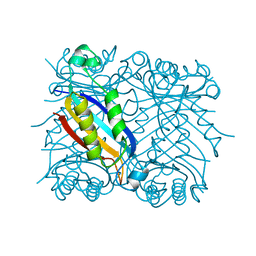 | | 1.65 ANGSTROM RESOLUTION STRUCTURE OF 7,8-DIHYDRONEOPTERIN ALDOLASE FROM STAPHYLOCOCCUS AUREUS | | Descriptor: | 7,8-DIHYDRONEOPTERIN ALDOLASE | | Authors: | Hennig, M, D'Arcy, A, Hampele, I.C, Page, M.G.P, Oefner, C.H, Dale, G. | | Deposit date: | 1998-03-31 | | Release date: | 1999-04-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure and reaction mechanism of 7,8-dihydroneopterin aldolase from Staphylococcus aureus.
Nat.Struct.Biol., 5, 1998
|
|
1CBK
 
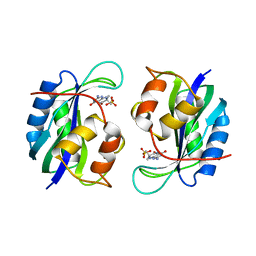 | | 7,8-DIHYDRO-6-HYDROXYMETHYLPTERIN-PYROPHOSPHOKINASE FROM HAEMOPHILUS INFLUENZAE | | Descriptor: | 7,8-DIHYDRO-7,7-DIMETHYL-6-HYDROXYPTERIN, PROTEIN (7,8-DIHYDRO-6-HYDROXYMETHYLPTERIN-PYROPHOSPHOKINASE), SULFATE ION | | Authors: | Hennig, M, D'Arcy, A, Dale, G, Oefner, C. | | Deposit date: | 1999-02-26 | | Release date: | 2000-03-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The structure and function of the 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase from Haemophilus influenzae.
J.Mol.Biol., 287, 1999
|
|
1IGS
 
 | | INDOLE-3-GLYCEROLPHOSPHATE SYNTHASE FROM SULFOLOBUS SOLFATARICUS AT 2.0 A RESOLUTION | | Descriptor: | INDOLE-3-GLYCEROLPHOSPHATE SYNTHASE, PHOSPHATE ION | | Authors: | Hennig, M, Darimont, B, Kirschner, K, Jansonius, J.N. | | Deposit date: | 1995-08-11 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0 A structure of indole-3-glycerol phosphate synthase from the hyperthermophile Sulfolobus solfataricus: possible determinants of protein stability.
Structure, 3, 1995
|
|
1LBM
 
 | |
1RWQ
 
 | | Human Dipeptidyl peptidase IV in complex with 5-aminomethyl-6-(2,4-dichloro-phenyl)-2-(3,5-dimethoxy-phenyl)-pyrimidin-4-ylamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-(AMINOMETHYL)-6-(2,4-DICHLOROPHENYL)-2-(3,5-DIMETHOXYPHENYL)PYRIMIDIN-4-AMINE, Dipeptidyl peptidase IV | | Authors: | Hennig, M, Thoma, R, Stihle, M. | | Deposit date: | 2003-12-17 | | Release date: | 2004-12-17 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Aminomethylpyrimidines as novel DPP-IV inhibitors: a 10(5)-fold activity increase by optimization of aromatic substituents
Bioorg.Med.Chem.Lett., 14, 2004
|
|
2BIT
 
 | | Crystal structure of human cyclophilin D at 1.7 A resolution | | Descriptor: | PEPTIDYL-PROLYL CIS-TRANS ISOMERASE | | Authors: | Hennig, M, Thoma, R, Stihle, M, Schlatter, D. | | Deposit date: | 2005-01-26 | | Release date: | 2005-01-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal Engineering Yields Crystals of Cyclophilin D Diffracting to 1.7 A Resolution
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2BIU
 
 | | Crystal structure of human cyclophilin D at 1.7 A resolution, DMSO complex | | Descriptor: | DIMETHYL SULFOXIDE, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE | | Authors: | Hennig, M, Thoma, R, Stihle, M, Schlatter, D. | | Deposit date: | 2005-01-26 | | Release date: | 2005-01-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal Engineering Yields Crystals of Cyclophilin D Diffracting to 1.7 A Resolution
Acta Crystallogr.,Sect.D, 61, 2005
|
|
3OC0
 
 | | Structure of human DPP-IV with HTS hit (2S,3S,11bS)-3-butyl-9,10-dimethoxy-1,3,4,6,7,11b-hexahydro-2H-pyrido[2,1-a]isoquinolin-2-ylamine | | Descriptor: | (2S,3R,11bR)-3-butyl-9,10-dimethoxy-1,3,4,6,7,11b-hexahydro-2H-pyrido[2,1-a]isoquinolin-2-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4 | | Authors: | Hennig, M, Stihle, M, Thoma, R. | | Deposit date: | 2010-08-09 | | Release date: | 2010-08-25 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of carmegliptin: A potent and long-acting dipeptidyl peptidase IV inhibitor for the treatment of type 2 diabetes.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3KWF
 
 | | human DPP-IV with carmegliptin (S)-1-((2S,3S,11bS)-2-Amino-9,10-dimethoxy-1,3,4,6,7,11b-hexahydro-2H-pyrido[2,1-a]isoquinolin-3-yl)-4-fluoromethyl-pyrrolidin-2-one | | Descriptor: | (4S)-1-[(2S,3S,11bS)-2-amino-9,10-dimethoxy-1,3,4,6,7,11b-hexahydro-2H-pyrido[2,1-a]isoquinolin-3-yl]-4-(fluoromethyl)pyrrolidin-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4 | | Authors: | Hennig, M, Stihle, M, Thoma, R. | | Deposit date: | 2009-12-01 | | Release date: | 2010-01-12 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of carmegliptin: A potent and long-acting dipeptidyl peptidase IV inhibitor for the treatment of type 2 diabetes.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3KWJ
 
 | | Structure of human DPP-IV with (2S,3S,11bS)-3-(3-Fluoromethyl-phenyl)-9,10-dimethoxy-1,3,4,6,7,11b-hexahydro-2H-pyrido[2,1-a]isoquinolin-2-ylamine | | Descriptor: | (2S,3S,11bS)-3-[3-(fluoromethyl)phenyl]-9,10-dimethoxy-1,3,4,6,7,11b-hexahydro-2H-pyrido[2,1-a]isoquinolin-2-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4 soluble form | | Authors: | Hennig, M, Stihle, M, Thoma, R. | | Deposit date: | 2009-12-01 | | Release date: | 2010-09-15 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Aryl- and heteroaryl-substituted aminobenzo[a]quinolizines as dipeptidyl peptidase IV inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
1NU6
 
 | | Crystal structure of human Dipeptidyl Peptidase IV (DPP-IV) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase IV, MERCURY (II) ION | | Authors: | Hennig, M, Stihle, M, Thoma, R, Ruf, A. | | Deposit date: | 2003-01-31 | | Release date: | 2003-08-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of Proline-Specific Exopeptidase Activity as Observed in Human Dipeptidyl Peptidase-IV.
Structure, 11, 2003
|
|
2JID
 
 | | Human Dipeptidyl peptidase IV in complex with 1-(3,4-Dimethoxy-phenyl) -3-m-tolyl-piperidine-4-ylamine | | Descriptor: | (3R,4S)-1-(3,4-DIMETHOXYPHENYL)-3-(3-METHYLPHENYL)PIPERIDIN-4-AMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIPEPTIDYL PEPTIDASE 4 | | Authors: | Hennig, M, Stihle, M, Luebbers, T, Thoma, R. | | Deposit date: | 2007-02-28 | | Release date: | 2008-05-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 1,3-Disubstituted 4-Aminopiperidines as Useful Tools in the Optimization of the 2-Aminobenzo[A]Quinolizine Dipeptidyl Peptidase Iv Inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
6ED9
 
 | |
3SGK
 
 | | Unique carbohydrate/carbohydrate interactions are required for high affinity binding of FcgIII and antibodies lacking core fucose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)][2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fc fragment, ... | | Authors: | Ferrara, C, Grau, S, Jaeger, C, Sondermann, P, Bruenker, P, Waldhauer, I, Hennig, M, Ruf, A, Rufer, A.C, Stihle, M, Umana, P, Benz, J. | | Deposit date: | 2011-06-15 | | Release date: | 2011-08-03 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Unique carbohydrate-carbohydrate interactions are required for high affinity binding between FcgammaRIII and antibodies lacking core fucose.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3SGJ
 
 | | Unique carbohydrate-carbohydrate interactions are required for high affinity binding between FcgIII and antibodies lacking core fucose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MALONATE ION, ... | | Authors: | Ferrara, C, Grau, S, Jaeger, C, Sondermann, P, Bruenker, P, Waldhauer, I, Hennig, M, Ruf, A, Rufer, A.C, Stihle, M, Umana, P, Benz, J. | | Deposit date: | 2011-06-15 | | Release date: | 2011-08-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unique carbohydrate-carbohydrate interactions are required for high affinity binding between FcgammaRIII and antibodies lacking core fucose.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
