1QI3
 
 | | MUTANT (D193N) MALTOTETRAOSE-FORMING EXO-AMYLASE IN COMPLEX WITH MALTOTETRAOSE | | Descriptor: | CALCIUM ION, PROTEIN (EXO-MALTOTETRAOHYDROLASE), alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Hasegawa, K, Kubota, M, Matsuura, Y. | | Deposit date: | 1999-06-01 | | Release date: | 1999-11-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Roles of catalytic residues in alpha-amylases as evidenced by the structures of the product-complexed mutants of a maltotetraose-forming amylase.
Protein Eng., 12, 1999
|
|
1QI5
 
 | | MUTANT (D294N) MALTOTETRAOSE-FORMING EXO-AMYLASE IN COMPLEX WITH MALTOTETRAOSE | | Descriptor: | CALCIUM ION, PROTEIN (EXO-MALTOTETRAOHYDROLASE), alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Hasegawa, K, Kubota, M, Matsuura, Y. | | Deposit date: | 1999-06-03 | | Release date: | 1999-11-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Roles of catalytic residues in alpha-amylases as evidenced by the structures of the product-complexed mutants of a maltotetraose-forming amylase.
Protein Eng., 12, 1999
|
|
1QI4
 
 | | MUTANT (E219G) MALTOTETRAOSE-FORMING EXO-AMYLASE IN COMPLEX WITH MALTOTETRAOSE | | Descriptor: | CALCIUM ION, PROTEIN (EXO-MALTOTETRAOHYDROLASE), alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Hasegawa, K, Kubota, M, Matsuura, Y. | | Deposit date: | 1999-06-01 | | Release date: | 1999-11-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Roles of catalytic residues in alpha-amylases as evidenced by the structures of the product-complexed mutants of a maltotetraose-forming amylase.
Protein Eng., 12, 1999
|
|
5GX3
 
 | | Luciferin-regenerating enzyme collected with serial synchrotron rotational crystallography with accumulated dose of 6.9 MGy (6th measurement) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, Luciferin regenerating enzyme, ... | | Authors: | Hasegawa, K, Yamashita, K, Murai, T, Nuemket, N, Hirata, K, Ueno, G, Ago, H, Nakatsu, T, Kumasaka, T, Yamamoto, M. | | Deposit date: | 2016-09-15 | | Release date: | 2017-01-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Development of a dose-limiting data collection strategy for serial synchrotron rotation crystallography
J Synchrotron Radiat, 24, 2017
|
|
5GX5
 
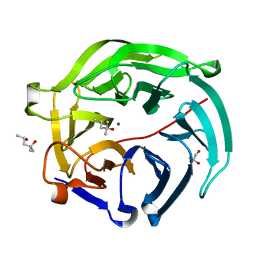 | | Luciferin-regenerating enzyme collected with serial synchrotron rotational crystallography with accumulated dose of 26 MGy (23rd measurement) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, Luciferin regenerating enzyme, ... | | Authors: | Hasegawa, K, Yamashita, K, Murai, T, Nuemket, N, Hirata, K, Ueno, G, Ago, H, Nakatsu, T, Kumasaka, T, Yamamoto, M. | | Deposit date: | 2016-09-15 | | Release date: | 2017-01-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Development of a dose-limiting data collection strategy for serial synchrotron rotation crystallography
J Synchrotron Radiat, 24, 2017
|
|
5GX1
 
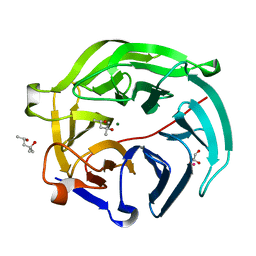 | | Luciferin-regenerating enzyme collected with serial synchrotron rotational crystallography with accumulated dose of 1.1 MGy (1st measurement) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, Luciferin regenerating enzyme, ... | | Authors: | Hasegawa, K, Yamashita, K, Murai, T, Nuemket, N, Hirata, K, Ueno, G, Ago, H, Nakatsu, T, Kumasaka, T, Yamamoto, M. | | Deposit date: | 2016-09-15 | | Release date: | 2017-01-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Development of a dose-limiting data collection strategy for serial synchrotron rotation crystallography
J Synchrotron Radiat, 24, 2017
|
|
5GX2
 
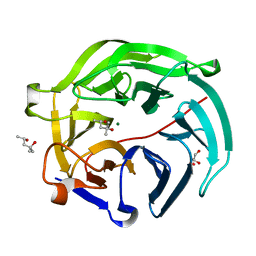 | | Luciferin-regenerating enzyme collected with serial synchrotron rotational crystallography with accumulated dose of 3.4 MGy (3rd measurement) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, Luciferin regenerating enzyme, ... | | Authors: | Hasegawa, K, Yamashita, K, Murai, T, Nuemket, N, Hirata, K, Ueno, G, Ago, H, Nakatsu, T, Kumasaka, T, Yamamoto, M. | | Deposit date: | 2016-09-15 | | Release date: | 2017-01-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Development of a dose-limiting data collection strategy for serial synchrotron rotation crystallography
J Synchrotron Radiat, 24, 2017
|
|
5GX4
 
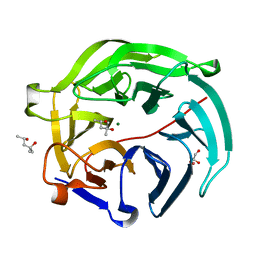 | | Luciferin-regenerating enzyme collected with serial synchrotron rotational crystallography with accumulated dose of 14 MGy (12th measurement) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, Luciferin regenerating enzyme, ... | | Authors: | Hasegawa, K, Yamashita, K, Murai, T, Nuemket, N, Hirata, K, Ueno, G, Ago, H, Nakatsu, T, Kumasaka, T, Yamamoto, M. | | Deposit date: | 2016-09-15 | | Release date: | 2017-01-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Development of a dose-limiting data collection strategy for serial synchrotron rotation crystallography
J Synchrotron Radiat, 24, 2017
|
|
7CDU
 
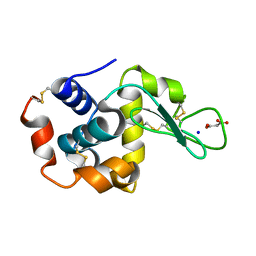 | | Lysozyme room-temperature structure determined by SS-ROX combined with HAG method, 1700 kGy (3000 images) | | Descriptor: | Lysozyme C, MALONATE ION, SODIUM ION | | Authors: | Hasegawa, K, Baba, S, Kawamura, T, Yamamoto, M, Kumasaka, T. | | Deposit date: | 2020-06-20 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evaluation of the data-collection strategy for room-temperature micro-crystallography studied by serial synchrotron rotation crystallography combined with the humid air and glue-coating method.
Acta Crystallogr.,Sect.D, 77, 2021
|
|
7CDT
 
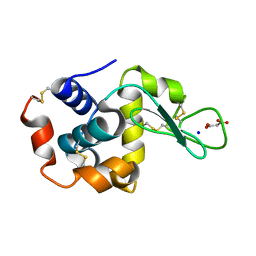 | | Lysozyme room-temperature structure determined by SS-ROX combined with HAG method, 830 kGy (3000 images) | | Descriptor: | Lysozyme C, MALONATE ION, SODIUM ION | | Authors: | Hasegawa, K, Baba, S, Kawamura, T, Yamamoto, M, Kumasaka, T. | | Deposit date: | 2020-06-20 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evaluation of the data-collection strategy for room-temperature micro-crystallography studied by serial synchrotron rotation crystallography combined with the humid air and glue-coating method.
Acta Crystallogr.,Sect.D, 77, 2021
|
|
7CDS
 
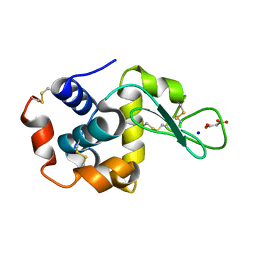 | | Lysozyme room-temperature structure determined by SS-ROX combined with HAG method, 420 kGy (3000 images) | | Descriptor: | Lysozyme C, MALONATE ION, SODIUM ION | | Authors: | Hasegawa, K, Baba, S, Kawamura, T, Yamamoto, M, Kumasaka, T. | | Deposit date: | 2020-06-20 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evaluation of the data-collection strategy for room-temperature micro-crystallography studied by serial synchrotron rotation crystallography combined with the humid air and glue-coating method.
Acta Crystallogr.,Sect.D, 77, 2021
|
|
7CDK
 
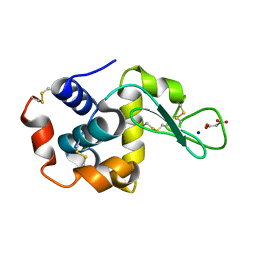 | | Lysozyme room-temperature structure determined by SS-ROX combined with HAG method, 42 kGy (4500 images from 1st half of data set) | | Descriptor: | Lysozyme C, MALONATE ION, SODIUM ION | | Authors: | Hasegawa, K, Baba, S, Kawamura, T, Yamamoto, M, Kumasaka, T. | | Deposit date: | 2020-06-20 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evaluation of the data-collection strategy for room-temperature micro-crystallography studied by serial synchrotron rotation crystallography combined with the humid air and glue-coating method.
Acta Crystallogr.,Sect.D, 77, 2021
|
|
7CDM
 
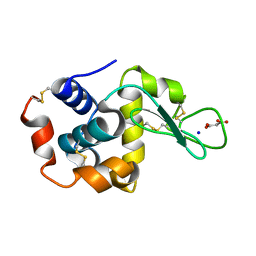 | | Lysozyme room-temperature structure determined by SS-ROX combined with HAG method, 42 kGy (4500 images from 2nd half of data set) | | Descriptor: | Lysozyme C, MALONATE ION, SODIUM ION | | Authors: | Hasegawa, K, Baba, S, Kawamura, T, Yamamoto, M, Kumasaka, T. | | Deposit date: | 2020-06-20 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evaluation of the data-collection strategy for room-temperature micro-crystallography studied by serial synchrotron rotation crystallography combined with the humid air and glue-coating method.
Acta Crystallogr.,Sect.D, 77, 2021
|
|
7CDN
 
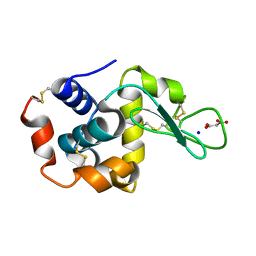 | | Lysozyme room-temperature structure determined by SS-ROX combined with HAG method, 42 kGy (9000 images) | | Descriptor: | Lysozyme C, MALONATE ION, SODIUM ION | | Authors: | Hasegawa, K, Baba, S, Kawamura, T, Yamamoto, M, Kumasaka, T. | | Deposit date: | 2020-06-20 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evaluation of the data-collection strategy for room-temperature micro-crystallography studied by serial synchrotron rotation crystallography combined with the humid air and glue-coating method.
Acta Crystallogr.,Sect.D, 77, 2021
|
|
7CDP
 
 | | Lysozyme room-temperature structure determined by SS-ROX combined with HAG method, 42 kGy (3000 images) | | Descriptor: | Lysozyme C, MALONATE ION, SODIUM ION | | Authors: | Hasegawa, K, Baba, S, Kawamura, T, Yamamoto, M, Kumasaka, T. | | Deposit date: | 2020-06-20 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evaluation of the data-collection strategy for room-temperature micro-crystallography studied by serial synchrotron rotation crystallography combined with the humid air and glue-coating method.
Acta Crystallogr.,Sect.D, 77, 2021
|
|
7CDQ
 
 | | Lysozyme room-temperature structure determined by SS-ROX combined with HAG method, 83 kGy (3000 images) | | Descriptor: | Lysozyme C, MALONATE ION, SODIUM ION | | Authors: | Hasegawa, K, Baba, S, Kawamura, T, Yamamoto, M, Kumasaka, T. | | Deposit date: | 2020-06-20 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evaluation of the data-collection strategy for room-temperature micro-crystallography studied by serial synchrotron rotation crystallography combined with the humid air and glue-coating method.
Acta Crystallogr.,Sect.D, 77, 2021
|
|
7CDR
 
 | | Lysozyme room-temperature structure determined by SS-ROX combined with HAG method, 210 kGy (3000 images) | | Descriptor: | Lysozyme C, MALONATE ION, SODIUM ION | | Authors: | Hasegawa, K, Baba, S, Kawamura, T, Yamamoto, M, Kumasaka, T. | | Deposit date: | 2020-06-20 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evaluation of the data-collection strategy for room-temperature micro-crystallography studied by serial synchrotron rotation crystallography combined with the humid air and glue-coating method.
Acta Crystallogr.,Sect.D, 77, 2021
|
|
7CDO
 
 | | Lysozyme room-temperature structure determined by SS-ROX combined with HAG method, 21 kGy (3000 images) | | Descriptor: | Lysozyme C, MALONATE ION, SODIUM ION | | Authors: | Hasegawa, K, Baba, S, Kawamura, T, Yamamoto, M, Kumasaka, T. | | Deposit date: | 2020-06-20 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evaluation of the data-collection strategy for room-temperature micro-crystallography studied by serial synchrotron rotation crystallography combined with the humid air and glue-coating method.
Acta Crystallogr.,Sect.D, 77, 2021
|
|
2E4M
 
 | | Crystal structure of hemagglutinin subcomponent complex (HA-33/HA-17) from Clostridium botulinum serotype D strain 4947 | | Descriptor: | HA-17, Main hemagglutinin component | | Authors: | Hasegawa, K, Watanabe, T, Suzuki, T, Yamano, A, Niwa, K, Ohyama, T. | | Deposit date: | 2006-12-13 | | Release date: | 2007-06-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Novel Subunit Structure of Clostridium botulinum Serotype D Toxin Complex with Three Extended Arms
J.Biol.Chem., 282, 2007
|
|
1QPK
 
 | | MUTANT (D193G) MALTOTETRAOSE-FORMING EXO-AMYLASE IN COMPLEX WITH MALTOTETRAOSE | | Descriptor: | CALCIUM ION, PROTEIN (MALTOTETRAOSE-FORMING AMYLASE), alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yoshioka, Y, Hasegawa, K, Matsuura, Y, Katsube, Y, Kubota, M. | | Deposit date: | 1999-05-26 | | Release date: | 1999-11-17 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Roles of catalytic residues in alpha-amylases as evidenced by the structures of the product-complexed mutants of a maltotetraose-forming amylase.
Protein Eng., 12, 1999
|
|
7X5N
 
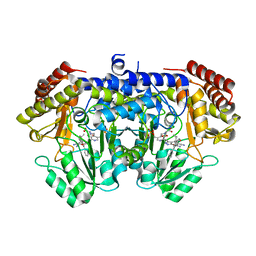 | | Crystal structure of E. faecium SHMT in complex with (+)-SHIN-1 and PLP-Ser | | Descriptor: | (4R)-6-azanyl-4-[3-(hydroxymethyl)-5-phenyl-phenyl]-3-methyl-4-propan-2-yl-1H-pyrano[2,3-c]pyrazole-5-carbonitrile, Serine hydroxymethyltransferase, [3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-SERINE | | Authors: | Hasegawa, K, Hayashi, H. | | Deposit date: | 2022-03-05 | | Release date: | 2022-07-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Serine hydroxymethyltransferase as a potential target of antibacterial agents acting synergistically with one-carbon metabolism-related inhibitors.
Commun Biol, 5, 2022
|
|
7X5O
 
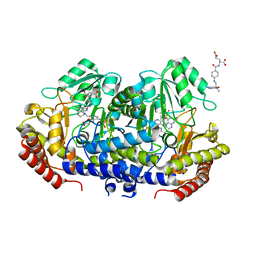 | | Crystal structure of E. faecium SHMT in complex with Me-THF and PLP-Gly | | Descriptor: | N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], N-[4-({[(6S)-2-AMINO-4-HYDROXY-5-METHYL-5,6,7,8-TETRAHYDROPTERIDIN-6-YL]METHYL}AMINO)BENZOYL]-L-GLUTAMIC ACID, Serine hydroxymethyltransferase | | Authors: | Hasegawa, K, Hayashi, H. | | Deposit date: | 2022-03-05 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Serine hydroxymethyltransferase as a potential target of antibacterial agents acting synergistically with one-carbon metabolism-related inhibitors.
Commun Biol, 5, 2022
|
|
7YVZ
 
 | | Structure of Caenorhabditis elegans CISD-1/mitoNEET | | Descriptor: | CDGSH iron-sulfur domain-containing protein 1 (CISD-1/mitoNEET), FE2/S2 (INORGANIC) CLUSTER | | Authors: | Hasegawa, K, Hagiuda, E, Taguchi, A.T, Geldenhuys, W, Iwasaki, T, Kumasaka, T. | | Deposit date: | 2022-08-20 | | Release date: | 2022-10-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and biological evaluation of Caenorhabditis elegans CISD-1/mitoNEET, a KLP-17 tail domain homologue, supports attenuation of paraquat-induced oxidative stress through a p38 MAPK-mediated antioxidant defense response
Advances in Redox Research, 6, 2022
|
|
6XYO
 
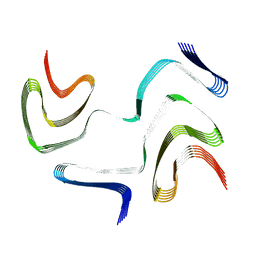 | | Multiple system atrophy Type I alpha-synuclein filament | | Descriptor: | Alpha-synuclein | | Authors: | Schweighauser, M, Shi, Y, Tarutani, A, Kametani, F, Murzin, A.G, Ghetti, B, Matsubara, T, Tomita, T, Ando, T, Hasegawa, K, Murayama, S, Yoshida, M, Hasegawa, M, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2020-01-30 | | Release date: | 2020-02-12 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structures of alpha-synuclein filaments from multiple system atrophy.
Nature, 585, 2020
|
|
6XYP
 
 | | Multiple system atrophy Type II-1 alpha-synuclein filament | | Descriptor: | Alpha-synuclein | | Authors: | Schweighauser, M, Shi, Y, Tarutani, A, Kametani, F, Murzin, A.G, Ghetti, B, Matsubara, T, Tomita, T, Ando, T, Hasegawa, K, Murayama, S, Yoshida, M, Hasegawa, M, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2020-01-30 | | Release date: | 2020-02-12 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structures of alpha-synuclein filaments from multiple system atrophy.
Nature, 585, 2020
|
|
