5XJL
 
 | |
8XGL
 
 | | Structure of the Magnaporthe oryzae effector NIS1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Necrosis-inducing secreted protein 1, ... | | Authors: | Han, R, Wang, D, Zhang, X, Liu, J. | | Deposit date: | 2023-12-15 | | Release date: | 2024-09-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Understanding and manipulating the recognition of necrosis-inducing secreted protein 1 (NIS1) by BRI1-associated receptor kinase 1 (BAK1).
Int.J.Biol.Macromol., 278, 2024
|
|
5DD1
 
 | | Crystal structures in an anti-HIV antibody lineage from immunization of Rhesus macaques | | Descriptor: | ANTI-HIV ANTIBODY DH570 FAB HEAVY CHAIN, ANTI-HIV ANTIBODY DH570 FAB LIGHT CHAIN | | Authors: | Zhang, R, Verkoczy, L, Wiehe, K, Alam, S.M, Nicely, N.I, Santra, S, Bradley, T, Pemble, C, Gao, F, Montefiori, D.C, Bouton-Verville, H, Kelsoe, G, Parks, R, Foulger, A, Tomaras, G, Keple, T.B, Moody, M.A, Liao, H.-X, Haynes, B.F. | | Deposit date: | 2015-08-24 | | Release date: | 2016-05-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.597 Å) | | Cite: | Initiation of immune tolerance-controlled HIV gp41 neutralizing B cell lineages.
Sci Transl Med, 8, 2016
|
|
5DD0
 
 | | Crystal structures in an anti-HIV antibody lineage from immunization of Rhesus macaques | | Descriptor: | ANTI-HIV ANTIBODY DH570 FAB HEAVY CHAIN, ANTI-HIV ANTIBODY DH570 FAB HEAVY LIGHT, oligo peptide | | Authors: | Zhang, R, Verkoczy, L, Wiehe, K, Alam, S.M, Nicely, N.I, Santra, S, Bradley, T, Pemble, C, Gao, F, Montefiori, D.C, Bouton-Verville, H, Kelsoe, G, Parks, R, Foulger, A, Tomaras, G, Keple, T.B, Moody, M.A, Liao, H.-X, Haynes, B.F. | | Deposit date: | 2015-08-24 | | Release date: | 2016-05-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.488 Å) | | Cite: | Initiation of immune tolerance-controlled HIV gp41 neutralizing B cell lineages.
Sci Transl Med, 8, 2016
|
|
5D68
 
 | | Crystal structure of KRIT1 ARD-FERM | | Descriptor: | Krev interaction trapped protein 1 | | Authors: | Zhang, R, Li, X, Boggon, T.J. | | Deposit date: | 2015-08-11 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.908 Å) | | Cite: | Structural analysis of the KRIT1 ankyrin repeat and FERM domains reveals a conformationally stable ARD-FERM interface.
J.Struct.Biol., 192, 2015
|
|
8U1T
 
 | | SARS-CoV-2 Envelope Protein Transmembrane Domain: Dimeric Structure Determined by Solid-State NMR | | Descriptor: | Envelope small membrane protein | | Authors: | Zhang, R, Qin, H, Prasad, R, Fu, R, Zhou, H.X, Cross, T. | | Deposit date: | 2023-09-02 | | Release date: | 2023-11-15 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Dimeric Transmembrane Structure of the SARS-CoV-2 E Protein.
Commun Biol, 6, 2023
|
|
3NZN
 
 | | The crystal structure of the Glutaredoxin from Methanosarcina mazei Go1 | | Descriptor: | GLYCEROL, Glutaredoxin, SULFATE ION | | Authors: | Zhang, R, Wu, R, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-16 | | Release date: | 2010-07-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The crystal structure of the Glutaredoxin from Methanosarcina mazei Go1
To be Published
|
|
9HMD
 
 | | Crystal structure of PDE6D in complex with DeltaTag (6a) | | Descriptor: | 4-[3,4-dimethyl-2-(4-methylphenyl)-7-oxidanylidene-pyrazolo[3,4-d]pyridazin-6-yl]-~{N}-[2-(3-ethylphenyl)ethyl]-~{N}-(piperidin-4-ylmethyl)butane-1-sulfonamide, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta | | Authors: | Zhang, R, Waldmann, H, Gasper, R. | | Deposit date: | 2024-12-09 | | Release date: | 2025-12-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of PDE6D in complex with DeltaTag (6a)
To be published
|
|
9HMC
 
 | | Crystal structure of PDE6D in complex with compound 5e | | Descriptor: | 4-[3,4-dimethyl-2-(4-methylphenyl)-7-oxidanylidene-pyrazolo[3,4-d]pyridazin-6-yl]-~{N}-heptyl-~{N}-(piperidin-4-ylmethyl)butane-1-sulfonamide, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Zhang, R, Waldmann, H, Gasper, R. | | Deposit date: | 2024-12-09 | | Release date: | 2025-12-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of PDE6D in complex with compound 5e
To be published
|
|
5LSH
 
 | | human lysozyme in complex with a tetrasaccharide fragment of the O-chain of LPS from Klebsiella pneumoniae | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION, ... | | Authors: | Zhang, R, Nifantiev, N.E, Krylov, V, Luetteke, T, Scheidig, A.J, Siebert, H.-C. | | Deposit date: | 2016-08-26 | | Release date: | 2017-06-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.061 Å) | | Cite: | Lysozyme's lectin-like characteristics facilitates its immune defense function.
Q. Rev. Biophys., 50, 2017
|
|
3CTM
 
 | | Crystal Structure of a Carbonyl Reductase from Candida Parapsilosis with anti-Prelog Stereo-specificity | | Descriptor: | Carbonyl Reductase | | Authors: | Zhang, R, Zhu, G, Li, X, Xu, Y, Zhang, X.C, Rao, Z. | | Deposit date: | 2008-04-14 | | Release date: | 2008-05-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystal structure of a carbonyl reductase from Candida parapsilosis with anti-Prelog stereospecificity.
Protein Sci., 17, 2008
|
|
8VP5
 
 | | Cryo-EM structure of the ABC transporter PCAT1 bound with ADP and Substrate | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 3-[oxidanyl-[2-(trimethyl-$l^{4}-azanyl)ethoxy]phosphoryl]oxypropyl hexadecanoate, ABC-type bacteriocin transporter, ... | | Authors: | Zhang, R, Jagessar, K.L, Polasa, A, Brownd, M, Stein, R, Moradi, M, Karakas, E, Mchaourab, H.S. | | Deposit date: | 2024-01-16 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Conformational cycle of a protease-containing ABC transporter in lipid nanodiscs reveals the mechanism of cargo-protein coupling.
Nat Commun, 15, 2024
|
|
8VPA
 
 | | Cryo-EM structure of the ABC transporter PCAT1 bound with ADP | | Descriptor: | ABC-type bacteriocin transporter, ADENOSINE-5'-DIPHOSPHATE | | Authors: | Zhang, R, Jagessar, K.L, Polasa, A, Brownd, M, Stein, R, Moradi, M, Karakas, E, Mchaourab, H.S. | | Deposit date: | 2024-01-16 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Conformational cycle of a protease-containing ABC transporter in lipid nanodiscs reveals the mechanism of cargo-protein coupling.
Nat Commun, 15, 2024
|
|
8VP9
 
 | | Cryo-EM structure of the cysteine-free ABC transporter PCAT1 bound with ADP and Substrate | | Descriptor: | 3-[oxidanyl-[2-(trimethyl-$l^{4}-azanyl)ethoxy]phosphoryl]oxypropyl hexadecanoate, ABC-type bacteriocin transporter, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Zhang, R, Jagessar, K.L, Polasa, A, Brownd, M, Stein, R, Moradi, M, Karakas, E, Mchaourab, H.S. | | Deposit date: | 2024-01-16 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Conformational cycle of a protease-containing ABC transporter in lipid nanodiscs reveals the mechanism of cargo-protein coupling.
Nat Commun, 15, 2024
|
|
8VOX
 
 | | Cryo-EM structure of the ABC transporter PCAT1 cysteine-free core bound with MgADP | | Descriptor: | ABC-type bacteriocin transporter, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Zhang, R, Jagessar, K.L, Polasa, A, Brownd, M, Stein, R, Moradi, M, Karakas, E, Mchaourab, H.S. | | Deposit date: | 2024-01-16 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Conformational cycle of a protease-containing ABC transporter in lipid nanodiscs reveals the mechanism of cargo-protein coupling.
Nat Commun, 15, 2024
|
|
8VP8
 
 | | Cryo-EM structure of the ABC transporter PCAT1 bound with ATP and Substrate | | Descriptor: | ABC-type bacteriocin transporter, ADENOSINE-5'-TRIPHOSPHATE | | Authors: | Zhang, R, Jagessar, K.L, Polasa, A, Brownd, M, Stein, R, Moradi, M, Karakas, E, Mchaourab, H.S. | | Deposit date: | 2024-01-16 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Conformational cycle of a protease-containing ABC transporter in lipid nanodiscs reveals the mechanism of cargo-protein coupling.
Nat Commun, 15, 2024
|
|
8VPB
 
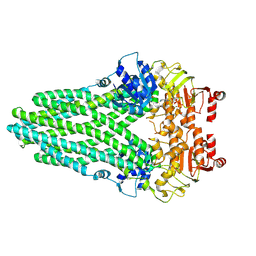 | | Cryo-EM structure of the ABC transporter PCAT1 bound with MgADP_Class_2 | | Descriptor: | ABC-type bacteriocin transporter, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Zhang, R, Jagessar, K.L, Polasa, A, Brownd, M, Stein, R, Moradi, M, Karakas, E, Mchaourab, H.S. | | Deposit date: | 2024-01-16 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Conformational cycle of a protease-containing ABC transporter in lipid nanodiscs reveals the mechanism of cargo-protein coupling.
Nat Commun, 15, 2024
|
|
8VP1
 
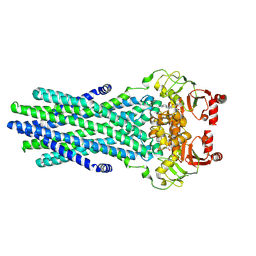 | | Cryo-EM structure of the ABC transporter PCAT1 cysteine-free core bound with ATP | | Descriptor: | ABC-type bacteriocin transporter, ADENOSINE-5'-TRIPHOSPHATE | | Authors: | Zhang, R, Jagessar, K.L, Polasa, A, Brownd, M, Stein, R, Moradi, M, Karakas, E, Mchaourab, H.S. | | Deposit date: | 2024-01-16 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Conformational cycle of a protease-containing ABC transporter in lipid nanodiscs reveals the mechanism of cargo-protein coupling.
Nat Commun, 15, 2024
|
|
8VOW
 
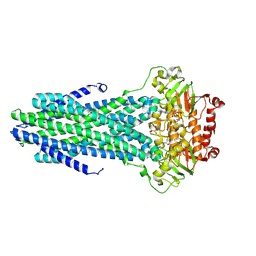 | | Cryo-EM structure of the ABC transporter PCAT1 cysteine-free core bound with MgATP and Vi | | Descriptor: | ABC-type bacteriocin transporter, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhang, R, Jagessar, K.L, Polasa, A, Brownd, M, Stein, R, Moradi, M, Karakas, E, Mchaourab, H.S. | | Deposit date: | 2024-01-16 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Conformational cycle of a protease-containing ABC transporter in lipid nanodiscs reveals the mechanism of cargo-protein coupling.
Nat Commun, 15, 2024
|
|
8VOZ
 
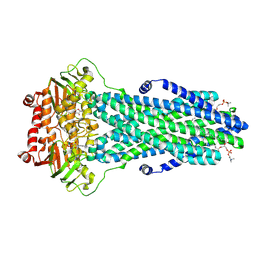 | | Cryo-EM structure of the ABC transporter PCAT1 cysteine-free core bound with MgADP and substrate | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 3-[oxidanyl-[2-(trimethyl-$l^{4}-azanyl)ethoxy]phosphoryl]oxypropyl hexadecanoate, ABC-type bacteriocin transporter, ... | | Authors: | Zhang, R, Jagessar, K.L, Polasa, A, Brownd, M, Stein, R, Moradi, M, Karakas, E, Mchaourab, H.S. | | Deposit date: | 2024-01-16 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Conformational cycle of a protease-containing ABC transporter in lipid nanodiscs reveals the mechanism of cargo-protein coupling.
Nat Commun, 15, 2024
|
|
8VP3
 
 | | Cryo-EM structure of the ABC transporter PCAT1 bound with MgADP and Substrate | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 3-[oxidanyl-[2-(trimethyl-$l^{4}-azanyl)ethoxy]phosphoryl]oxypropyl hexadecanoate, ABC-type bacteriocin transporter, ... | | Authors: | Zhang, R, Jagessar, K.L, Polasa, A, Brownd, M, Stein, R, Moradi, M, Karakas, E, Mchaourab, H.S. | | Deposit date: | 2024-01-16 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Conformational cycle of a protease-containing ABC transporter in lipid nanodiscs reveals the mechanism of cargo-protein coupling.
Nat Commun, 15, 2024
|
|
8VP6
 
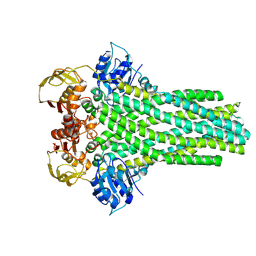 | | Cryo-EM structure of the ABC transporter PCAT1 bound with Mg_class_2 | | Descriptor: | ABC-type bacteriocin transporter | | Authors: | Zhang, R, Jagessar, K.L, Polasa, A, Brownd, M, Stein, R, Moradi, M, Karakas, E, Mchaourab, H.S. | | Deposit date: | 2024-01-16 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Conformational cycle of a protease-containing ABC transporter in lipid nanodiscs reveals the mechanism of cargo-protein coupling.
Nat Commun, 15, 2024
|
|
2OTD
 
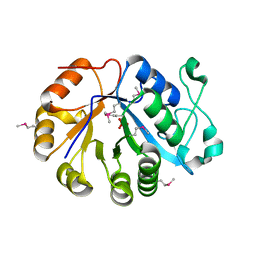 | | The crystal structure of the glycerophosphodiester phosphodiesterase from Shigella flexneri 2a | | Descriptor: | Glycerophosphodiester phosphodiesterase, PHOSPHATE ION | | Authors: | Zhang, R, Wu, R, Clancy, S, Jiang, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-02-07 | | Release date: | 2007-03-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of the glycerophosphodiester phosphodiesterase from Shigella flexneri 2a
To be Published
|
|
3O12
 
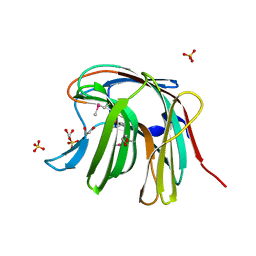 | | The crystal structure of a functionally unknown protein from Saccharomyces cerevisiae. | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Uncharacterized protein YJL217W | | Authors: | Zhang, R, Tan, K, Xu, X, Cui, H, Chin, S, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-20 | | Release date: | 2010-09-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of a functionally unknown protein from Saccharomyces cerevisiae.
TO BE PUBLISHED
|
|
3O2I
 
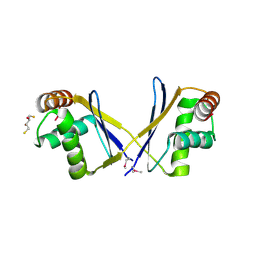 | | The crystal structure of a functionally unknown protein from Leptospirillum sp. Group II UBA | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, DI(HYDROXYETHYL)ETHER, Uncharacterized protein | | Authors: | Zhang, R, Tan, K, Xu, X, Cui, H, Ng, J, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-22 | | Release date: | 2010-09-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | The crystal structure of a functionally unknown protein from Leptospirillum sp. Group II UBA
TO BE PUBLISHED
|
|
