3S0Z
 
 | | Crystal structure of New Delhi Metallo-beta-lactamase (NDM-1) | | Descriptor: | Metallo-beta-lactamase, ZINC ION | | Authors: | Guo, Y, Wang, J, Niu, G.J, Shui, W.Q, Sun, Y.N, Lou, Z.Y, Rao, Z.H. | | Deposit date: | 2011-05-13 | | Release date: | 2011-06-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A structural view of the antibiotic degradation enzyme NDM-1 from a superbug.
Protein Cell, 2011
|
|
4LHO
 
 | | Crystal Structure of FG41Malonate Semialdehyde Decarboxylase inhibited by 3-bromopropiolate | | Descriptor: | 3-chloro-3-oxopropanoic acid, FG41 Malonate Semialdehyde Decarboxylase, PHOSPHATE ION | | Authors: | Guo, Y, Serrano, H, Poelarends, G.J, Johnson Jr, W.H, Hackert, M.L, Whitman, C.P. | | Deposit date: | 2013-07-01 | | Release date: | 2013-07-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.224 Å) | | Cite: | Kinetic, Mutational, and Structural Analysis of Malonate Semialdehyde Decarboxylase from Coryneform Bacterium Strain FG41: Mechanistic Implications for the Decarboxylase and Hydratase Activities.
Biochemistry, 52, 2013
|
|
8GOU
 
 | | Omicron BA.4/5 SARS-CoV-2 S in complex with TH003 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, TH003 Fab heavy chain, ... | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-25 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
6IWF
 
 | | Crystal structure of HitA from Pseudomonas aeruginosa | | Descriptor: | Ferric iron-binding protein HitA | | Authors: | Guo, Y, Zhengrui, Z, Li, H. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.70662332 Å) | | Cite: | Identification and Characterization of a Metalloprotein Involved in Gallium Internalization in Pseudomonas aeruginosa.
Acs Infect Dis., 5, 2019
|
|
7QD7
 
 | | TarM(Se)_G117R | | Descriptor: | CHLORIDE ION, GLYCEROL, PENTAETHYLENE GLYCOL, ... | | Authors: | Guo, Y, Stehle, T. | | Deposit date: | 2021-11-26 | | Release date: | 2023-05-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Invasive Staphylococcus epidermidis uses a unique processive wall teichoic acid glycosyltransferase to evade immune recognition.
Sci Adv, 9, 2023
|
|
7QH9
 
 | | TarM(Se)_G117R-4RboP | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, TarM(Se)_G117R-4RboP, ... | | Authors: | Guo, Y, Stehle, T. | | Deposit date: | 2021-12-10 | | Release date: | 2023-05-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.689 Å) | | Cite: | Invasive Staphylococcus epidermidis uses a unique processive wall teichoic acid glycosyltransferase to evade immune recognition.
Sci Adv, 9, 2023
|
|
7QNT
 
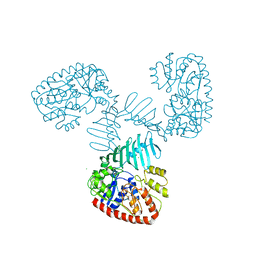 | | TarM(Se) native | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Guo, Y, Stehle, T. | | Deposit date: | 2021-12-22 | | Release date: | 2023-05-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Invasive Staphylococcus epidermidis uses a unique processive wall teichoic acid glycosyltransferase to evade immune recognition.
Sci Adv, 9, 2023
|
|
1FBM
 
 | | ASSEMBLY DOMAIN OF CARTILAGE OLIGOMERIC MATRIX PROTEIN IN COMPLEX WITH ALL-TRANS RETINOL | | Descriptor: | PROTEIN (CARTILAGE OLIGOMERIC MATRIX PROTEIN), RETINOL | | Authors: | Guo, Y, Bozic, D, Malashkevich, V.N, Kammerer, R.A, Schulthess, T. | | Deposit date: | 2000-07-16 | | Release date: | 2000-08-02 | | Last modified: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | All-trans retinol, vitamin D and other hydrophobic compounds bind in the axial pore of the five-stranded coiled-coil domain of cartilage oligomeric matrix protein.
EMBO J., 17, 1998
|
|
7L97
 
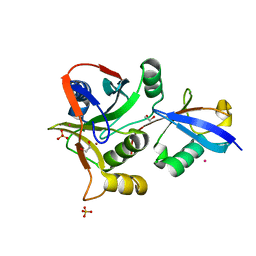 | | Crystal structure of STAMBPL1 in complex with an engineered binder | | Descriptor: | 1,2-ETHANEDIOL, AMSH-like protease, SULFATE ION, ... | | Authors: | Guo, Y, Dong, A, Hou, F, Li, Y, Zhang, W, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-01-02 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural and functional characterization of ubiquitin variant inhibitors for the JAMM-family deubiquitinases STAMBP and STAMBPL1.
J.Biol.Chem., 297, 2021
|
|
3MF8
 
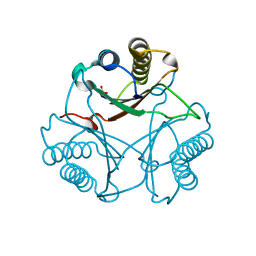 | | Crystal Structure of Native cis-CaaD | | Descriptor: | Cis-3-chloroacrylic acid dehalogenase, SULFATE ION | | Authors: | Guo, Y, Serrano, H, Ernst, S.R, Johnson Jr, W.H, Hackert, M.L, Whitman, C.P. | | Deposit date: | 2010-04-01 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structures of native and inactivated cis-3-chloroacrylic acid dehalogenase: Implications for the catalytic and inactivation mechanisms.
Bioorg.Chem., 39, 2011
|
|
3MF7
 
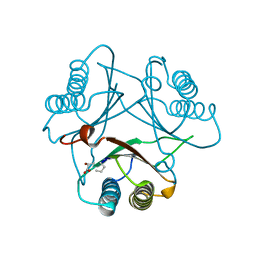 | | Crystal Structure of (R)-oxirane-2-carboxylate inhibited cis-CaaD | | Descriptor: | Cis-3-chloroacrylic acid dehalogenase | | Authors: | Guo, Y, Serrano, H, Ernst, S.R, Johnson Jr, W.H, Hackert, M.L, Whitman, C.P. | | Deposit date: | 2010-04-01 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of native and inactivated cis-3-chloroacrylic acid dehalogenase: Implications for the catalytic and inactivation mechanisms.
Bioorg.Chem., 39, 2011
|
|
3MQ3
 
 | |
3MJZ
 
 | | The crystal structure of native FG41 MSAD | | Descriptor: | FG41 Malonate Semialdehyde Decarboxylase | | Authors: | Guo, Y, Serrano, H, Poelarends, G.J, Johnson, W.H.Jr, Hackert, M.L, Whitman, C.P. | | Deposit date: | 2010-04-13 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Kinetic, Mutational, and Structural Analysis of Malonate Semialdehyde Decarboxylase from Coryneform Bacterium Strain FG41: Mechanistic Implications for the Decarboxylase and Hydratase Activities.
Biochemistry, 52, 2013
|
|
3MLC
 
 | | Crystal structure of FG41MSAD inactivated by 3-chloropropiolate | | Descriptor: | 3-chloro-3-oxopropanoic acid, FG41 Malonate Semialdehyde Decarboxylase | | Authors: | Guo, Y, Serrano, H, Poelarends, G.J, Johnson Jr, W.H, Hackert, M.L, Whitman, C.P. | | Deposit date: | 2010-04-16 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.224 Å) | | Cite: | Kinetic, Mutational, and Structural Analysis of Malonate Semialdehyde Decarboxylase from Coryneform Bacterium Strain FG41: Mechanistic Implications for the Decarboxylase and Hydratase Activities.
Biochemistry, 52, 2013
|
|
1SL5
 
 | | Crystal Structure of DC-SIGN carbohydrate recognition domain complexed with LNFP III (Dextra L504). | | Descriptor: | CALCIUM ION, MAGNESIUM ION, alpha-L-fucopyranose-(1-3)-[beta-D-galactopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose, ... | | Authors: | Guo, Y, Feinberg, H, Conroy, E, Mitchell, D.A, Alvarez, R, Blixt, O, Taylor, M.E, Weis, W.I, Drickamer, K. | | Deposit date: | 2004-03-05 | | Release date: | 2004-06-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for distinct ligand-binding and targeting properties of the receptors
DC-SIGN and DC-SIGNR
Nat.Struct.Mol.Biol., 11, 2004
|
|
1SL4
 
 | | Crystal Structure of DC-SIGN carbohydrate recognition domain complexed with Man4 | | Descriptor: | CALCIUM ION, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose, mDC-SIGN1B type I isoform | | Authors: | Guo, Y, Feinberg, H, Conroy, E, Mitchell, D.A, Alvarez, R, Blixt, O, Taylor, M.E, Weis, W.I, Drickamer, K. | | Deposit date: | 2004-03-05 | | Release date: | 2004-06-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for distinct ligand-binding and targeting properties of the receptors
DC-SIGN and DC-SIGNR
Nat.Struct.Mol.Biol., 11, 2004
|
|
1SL6
 
 | | Crystal Structure of a fragment of DC-SIGNR (containg the carbohydrate recognition domain and two repeats of the neck) complexed with Lewis-x. | | Descriptor: | C-type lectin DC-SIGNR, CALCIUM ION, alpha-L-fucopyranose-(1-3)-[beta-D-galactopyranose-(1-4)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Guo, Y, Feinberg, H, Conroy, E, Mitchell, D.A, Alvarez, R, Blixt, O, Taylor, M.E, Weis, W.I, Drickamer, K. | | Deposit date: | 2004-03-05 | | Release date: | 2004-06-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for distinct ligand-binding and targeting properties of the receptors
DC-SIGN and DC-SIGNR
Nat.Struct.Mol.Biol., 11, 2004
|
|
6V0T
 
 | | Crystal Structure of Catalytic Subunit of Bovine Pyruvate Dehydrogenase Phosphatase 1 - Catalytic Domain | | Descriptor: | MANGANESE (II) ION, SULFATE ION, [Pyruvate dehydrogenase [acetyl-transferring]]-phosphatase 1, ... | | Authors: | Guo, Y, Qiu, W, Ernst, S.R, Carroll, D.W, Hackert, M.L. | | Deposit date: | 2019-11-19 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the catalytic subunit of bovine pyruvate dehydrogenase phosphatase.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
8F6C
 
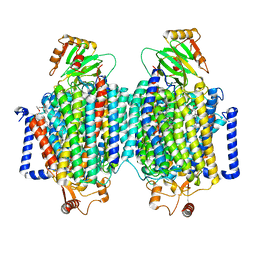 | | E. coli cytochrome bo3 ubiquinol oxidase dimer | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | Authors: | Guo, Y, Karimullina, E, Borek, D, Savchenko, A. | | Deposit date: | 2022-11-16 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Monomer and dimer structures of cytochrome bo 3 ubiquinol oxidase from Escherichia coli.
Protein Sci., 32, 2023
|
|
8F68
 
 | | E. coli cytochrome bo3 ubiquinol oxidase monomer | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | Authors: | Guo, Y, Karimullina, E, Borek, D, Savchenko, A. | | Deposit date: | 2022-11-16 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Monomer and dimer structures of cytochrome bo 3 ubiquinol oxidase from Escherichia coli.
Protein Sci., 32, 2023
|
|
5E04
 
 | | Crystal structure of Andes virus nucleoprotein | | Descriptor: | Nucleoprotein | | Authors: | Guo, Y, Wang, W.M, Lou, Z.Y. | | Deposit date: | 2015-09-28 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of the Core Region of Hantavirus Nucleocapsid Protein Reveals the Mechanism for Ribonucleoprotein Complex Formation
J.Virol., 90, 2015
|
|
5E06
 
 | |
5E05
 
 | |
4RYQ
 
 | | Crystal structure of BcTSPO, type 2 at 1.7 Angstrom | | Descriptor: | Integral membrane protein, [(Z)-octadec-9-enyl] (2R)-2,3-bis(oxidanyl)propanoate | | Authors: | Guo, Y, Liu, Q, Hendrickson, W.A, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2014-12-16 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Protein structure. Structure and activity of tryptophan-rich TSPO proteins.
Science, 347, 2015
|
|
4RYM
 
 | |
