1EIY
 
 | | THE CRYSTAL STRUCTURE OF PHENYLALANYL-TRNA SYNTHETASE FROM THERMUS THERMOPHILUS COMPLEXED WITH COGNATE TRNAPHE | | Descriptor: | PHENYLALANYL-TRNA SYNTHETASE, TRNA(PHE) | | Authors: | Goldgur, Y, Mosyak, L, Reshetnikova, L, Ankilova, V, Safro, M. | | Deposit date: | 2000-02-29 | | Release date: | 2000-03-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The crystal structure of phenylalanyl-tRNA synthetase from thermus thermophilus complexed with cognate tRNAPhe.
Structure, 5, 1997
|
|
1QS4
 
 | | Core domain of HIV-1 integrase complexed with Mg++ and 1-(5-chloroindol-3-yl)-3-hydroxy-3-(2H-tetrazol-5-yl)-propenone | | Descriptor: | 1-(5-CHLOROINDOL-3-YL)-3-HYDROXY-3-(2H-TETRAZOL-5-YL)-PROPENONE, MAGNESIUM ION, PROTEIN (HIV-1 INTEGRASE (E.C.2.7.7.49)) | | Authors: | Goldgur, Y, Craigie, R, Fujiwara, T, Yoshinaga, T, Davies, D.R. | | Deposit date: | 1999-06-25 | | Release date: | 1999-11-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the HIV-1 integrase catalytic domain complexed with an inhibitor: a platform for antiviral drug design.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1BIS
 
 | | HIV-1 INTEGRASE CORE DOMAIN | | Descriptor: | HIV-1 INTEGRASE | | Authors: | Goldgur, Y, Dyda, F, Hickman, A.B, Jenkins, T.M, Craigie, R, Davies, D.R. | | Deposit date: | 1998-06-19 | | Release date: | 1998-08-19 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1BIU
 
 | | HIV-1 INTEGRASE CORE DOMAIN COMPLEXED WITH MG++ | | Descriptor: | HIV-1 INTEGRASE, MAGNESIUM ION | | Authors: | Goldgur, Y, Dyda, F, Hickman, A.B, Jenkins, T.M, Craigie, R, Davies, D.R. | | Deposit date: | 1998-06-19 | | Release date: | 1998-08-19 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1BIZ
 
 | | HIV-1 INTEGRASE CORE DOMAIN | | Descriptor: | CACODYLATE ION, HIV-1 INTEGRASE | | Authors: | Goldgur, Y, Dyda, F, Hickman, A.B, Jenkins, T.M, Craigie, R, Davies, D.R. | | Deposit date: | 1998-06-21 | | Release date: | 1998-08-19 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
3ETP
 
 | | The crystal structure of the ligand-binding domain of the EphB2 receptor at 2.0 A resolution | | Descriptor: | Ephrin type-B receptor 2 | | Authors: | Goldgur, Y, Paavilainen, S, Nikolov, D.B, Himanen, J.P. | | Deposit date: | 2008-10-08 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the ligand-binding domain of the EphB2 receptor at 2 A resolution.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
4W4H
 
 | | Escherichia coli tryptophanase in holo form | | Descriptor: | Tryptophanase | | Authors: | Goldgur, Y. | | Deposit date: | 2014-08-14 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structures of Escherichia coli tryptophanase in holo and `semi-holo' forms.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4W1Y
 
 | |
2OQX
 
 | | Crystal Structure of the apo form of E. coli tryptophanase at 1.9 A resolution | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Goldgur, Y, Kogan, A, Gdalevsky, G, Parola, A, Cohen-Luria, R, Almog, O. | | Deposit date: | 2007-02-01 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of apo tryptophanase from Escherichia coli reveals a wide-open conformation.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
6DBP
 
 | |
6NVP
 
 | | Crystal structure of Pseudomonas putida nuclease MPE | | Descriptor: | MANGANESE (II) ION, Nuclease MPE | | Authors: | Goldgur, Y, Shuman, S, Ejaz, A. | | Deposit date: | 2019-02-05 | | Release date: | 2019-03-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Activity and structure ofPseudomonas putidaMPE, a manganese-dependent single-strand DNA endonuclease encoded in a nucleic acid repair gene cluster.
J.Biol.Chem., 294, 2019
|
|
6NVO
 
 | | Crystal structure of Pseudomonas putida nuclease MPE | | Descriptor: | MANGANESE (II) ION, Nuclease MPE | | Authors: | Goldgur, Y, Shuman, S, Ejaz, A. | | Deposit date: | 2019-02-05 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Activity and structure ofPseudomonas putidaMPE, a manganese-dependent single-strand DNA endonuclease encoded in a nucleic acid repair gene cluster.
J.Biol.Chem., 294, 2019
|
|
5TT6
 
 | | T4 RNA Ligase 1 (K99M) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, T4 RNA ligase 1 | | Authors: | Goldgur, Y, Unciuleac, M.-C, Shuman, S.H. | | Deposit date: | 2016-11-01 | | Release date: | 2017-03-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | Two-metal versus one-metal mechanisms of lysine adenylylation by ATP-dependent and NAD(+)-dependent polynucleotide ligases.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5TT5
 
 | | Escherichia coli LigA (K115M) in complex with NAD+ | | Descriptor: | DNA ligase, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Goldgur, Y, Unciuleac, M.-C, Shuman, S.H. | | Deposit date: | 2016-11-01 | | Release date: | 2017-03-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.552 Å) | | Cite: | Two-metal versus one-metal mechanisms of lysine adenylylation by ATP-dependent and NAD(+)-dependent polynucleotide ligases.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8E1H
 
 | | Asp1 kinase in complex with ADP Mg 5-IP7 | | Descriptor: | (1r,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, ADENOSINE-5'-DIPHOSPHATE, Inositol hexakisphosphate and diphosphoinositol-pentakisphosphate kinase, ... | | Authors: | Goldgur, Y, Shuman, S, Benjamin, B. | | Deposit date: | 2022-08-10 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Fission Yeast Inositol Pyrophosphate Kinase Asp1 in Ligand-Free, Substrate-Bound, and Product-Bound States.
Mbio, 13, 2022
|
|
8E1S
 
 | | Asp1 kinase in complex with ADPNP Mn IP6 | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Inositol hexakisphosphate and diphosphoinositol-pentakisphosphate kinase, MANGANESE (II) ION, ... | | Authors: | Goldgur, Y, Shuman, S, Benjamin, B. | | Deposit date: | 2022-08-11 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structures of Fission Yeast Inositol Pyrophosphate Kinase Asp1 in Ligand-Free, Substrate-Bound, and Product-Bound States.
Mbio, 13, 2022
|
|
8E1V
 
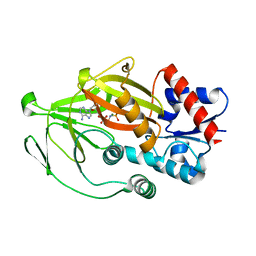 | | Asp1 kinase in complex with ADPNP Mg IP6 | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Inositol hexakisphosphate and diphosphoinositol-pentakisphosphate kinase, MAGNESIUM ION, ... | | Authors: | Goldgur, Y, Shuman, S, Benjamin, B. | | Deposit date: | 2022-08-11 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Fission Yeast Inositol Pyrophosphate Kinase Asp1 in Ligand-Free, Substrate-Bound, and Product-Bound States.
Mbio, 13, 2022
|
|
8E1I
 
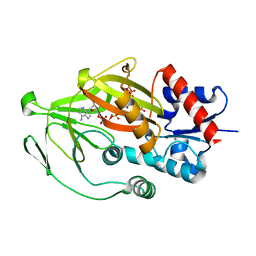 | | Asp1 kinase in complex with ATP Mg 5-IP7 | | Descriptor: | (1r,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, ADENOSINE-5'-TRIPHOSPHATE, Inositol hexakisphosphate and diphosphoinositol-pentakisphosphate kinase, ... | | Authors: | Goldgur, Y, Shuman, S, Benjamin, B. | | Deposit date: | 2022-08-10 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Fission Yeast Inositol Pyrophosphate Kinase Asp1 in Ligand-Free, Substrate-Bound, and Product-Bound States.
Mbio, 13, 2022
|
|
8E1J
 
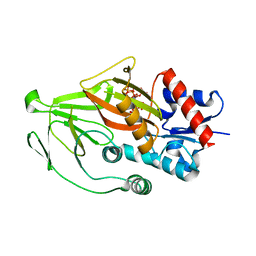 | | Asp1 kinase in complex with 1,5-IP8 | | Descriptor: | (1R,3S,4R,5S,6R)-2,4,5,6-tetrakis(phosphonooxy)cyclohexane-1,3-diyl bis[trihydrogen (diphosphate)], Inositol hexakisphosphate and diphosphoinositol-pentakisphosphate kinase | | Authors: | Goldgur, Y, Shuman, S, Benjamin, B. | | Deposit date: | 2022-08-10 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of Fission Yeast Inositol Pyrophosphate Kinase Asp1 in Ligand-Free, Substrate-Bound, and Product-Bound States.
Mbio, 13, 2022
|
|
8E1T
 
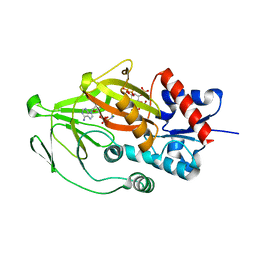 | | Asp1 kinase in complex with ADPNP Mg IP7 | | Descriptor: | (1r,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, Inositol hexakisphosphate and diphosphoinositol-pentakisphosphate kinase, MAGNESIUM ION, ... | | Authors: | Goldgur, Y, Shuman, S, Benjamin, B. | | Deposit date: | 2022-08-11 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structures of Fission Yeast Inositol Pyrophosphate Kinase Asp1 in Ligand-Free, Substrate-Bound, and Product-Bound States.
Mbio, 13, 2022
|
|
8F8V
 
 | | Crystal structure of Nb.X0 | | Descriptor: | Nb.X0 | | Authors: | Goldgur, Y, Ravetch, J, Gupta, A, Kao, K, Andi, B. | | Deposit date: | 2022-11-22 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Mechanism of glycoform specificity and in vivo protection by an anti-afucosylated IgG nanobody.
Nat Commun, 14, 2023
|
|
8F8W
 
 | | Crystal structure of Nb.X0 bound to the afucosylated human IgG1 fragment crystal form I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nb.X0, afucosylated IgG1 fragment | | Authors: | Goldgur, Y, Ravetch, J, Gupta, A, Kao, K, Oren, D. | | Deposit date: | 2022-11-22 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Mechanism of glycoform specificity and in vivo protection by an anti-afucosylated IgG nanobody.
Nat Commun, 14, 2023
|
|
8F8X
 
 | | Crystal structure of Nb.X0 bound to the afucosylated human IgG1 fragment crystal form II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nb.X0, Uncharacterized protein DKFZp686C11235 | | Authors: | Goldgur, Y, Ravetch, J, Gupta, A, Kao, K, Oren, D. | | Deposit date: | 2022-11-22 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanism of glycoform specificity and in vivo protection by an anti-afucosylated IgG nanobody.
Nat Commun, 14, 2023
|
|
7K7J
 
 | | EphB6 receptor ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ephrin type-B receptor 6 | | Authors: | Goldgur, Y, Himanen, J. | | Deposit date: | 2020-09-22 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | Structure of the EphB6 receptor ectodomain.
Plos One, 16, 2021
|
|
7LFQ
 
 | | Pyrococcus RNA ligase | | Descriptor: | DNA (5'-D(*AP*TP*GP*TP*CP*C)-3'), POTASSIUM ION, RNA-splicing ligase RtcB, ... | | Authors: | Goldgur, Y, Shuman, S, Banerjee, A. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of 3'-PO 4 /5'-OH RNA ligase RtcB in complex with a 5'-OH oligonucleotide.
Rna, 27, 2021
|
|
