1OH1
 
 | | Solution structure of staphostatin A form Staphylococcus aureus confirms the discovery of a novel class of cysteine proteinase inhibitors. | | Descriptor: | STAPHOSTATIN A | | Authors: | Dubin, G, Popowicz, G, Krajewski, M, Stec, J, Bochtler, M, Potempa, J, Dubin, A, Holak, T.A. | | Deposit date: | 2003-05-21 | | Release date: | 2003-11-20 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | A Novel Class of Cysteine Protease Inhibitors: Solution Structure of Staphostatin a from Staphylococcus Aureus
Biochemistry, 42, 2003
|
|
2VID
 
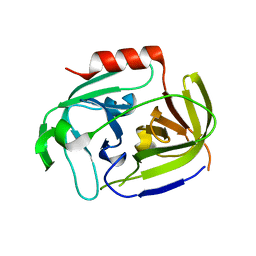 | | Serine protease SplB from Staphylococcus aureus at 1.8A resolution | | Descriptor: | SERINE PROTEASE SPLB | | Authors: | Dubin, G, Stec-Niemczyk, J, Kisielewska, M, Pustelny, K, Popowicz, G.M, Bista, M, Kantyka, T, Boulware, K.T, Stennicke, H.R, Czarna, A, Phopaisarn, M, Daugherty, P.S, Thogersen, I.B, Enghild, J.J, Thornberry, N, Dubin, A, Potempa, J. | | Deposit date: | 2007-11-30 | | Release date: | 2008-05-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Enzymatic Activity of the Staphylococcus Aureus Splb Serine Protease is Induced by Substrates Containing the Sequence Trp-Glu-Leu-Gln.
J.Mol.Biol., 379, 2008
|
|
5O12
 
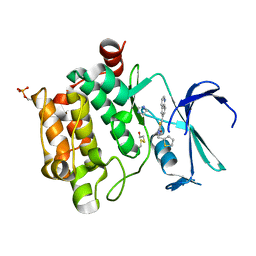 | |
5O13
 
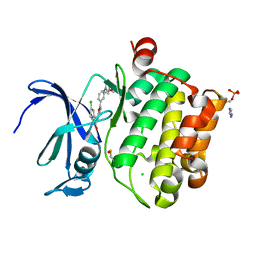 | | Crystal structure of PIM1 kinase in complex with small-molecule inhibitor | | Descriptor: | (3~{E})-5-chloranyl-3-[[5-[3-[(4-methyl-1,4-diazepan-1-yl)carbonyl]phenyl]furan-2-yl]methylidene]-1~{H}-indol-2-one, CHLORIDE ION, IMIDAZOLE, ... | | Authors: | Dubin, G, Bogusz, J. | | Deposit date: | 2017-05-17 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural analysis of PIM1 kinase complexes with ATP-competitive inhibitors.
Sci Rep, 7, 2017
|
|
4MVN
 
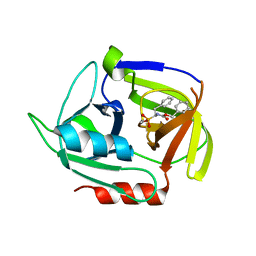 | | Crystal structure of the staphylococcal serine protease SplA in complex with a specific phosphonate inhibitor | | Descriptor: | Serine protease splA, [(1S)-1-{[(benzyloxy)carbonyl]amino}-2-phenylethyl]phosphonic acid | | Authors: | Zdzalik, M, Burchacka, E, Niemczyk, J.S, Pustelny, K, Popowicz, G.M, Wladyka, B, Dubin, A, Potempa, J, Sienczyk, M, Dubin, G, Oleksyszyn, J. | | Deposit date: | 2013-09-24 | | Release date: | 2014-01-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Development and binding characteristics of phosphonate inhibitors of SplA protease from Staphylococcus aureus.
Protein Sci., 23, 2014
|
|
3UFA
 
 | | Crystal structure of the staphylococcal serine protease SplA in complex with a specific phosphonate inhibitor | | Descriptor: | CHLORIDE ION, N-(3-carboxypropanoyl)-L-valyl-N-[(1S)-2-phenyl-1-phosphonoethyl]-L-prolinamide, Serine protease splA | | Authors: | Zdzalik, M, Pietrusewicz, E, Pustelny, K, Stec-Niemczyk, J, Popowicz, G.M, Potempa, J, Oleksyszyn, J, Dubin, G. | | Deposit date: | 2011-10-31 | | Release date: | 2013-01-23 | | Last modified: | 2014-03-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development and binding characteristics of phosphonate inhibitors of SplA protease from Staphylococcus aureus.
Protein Sci., 23, 2014
|
|
2AS9
 
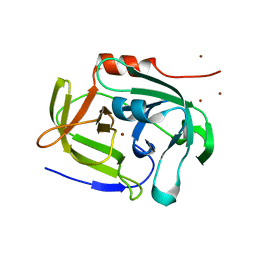 | | Functional and structural characterization of Spl proteases from staphylococcus aureus | | Descriptor: | ZINC ION, serine protease | | Authors: | Popowicz, G.M, Dubin, G, Stec-Niemczyk, J, Czarny, A, Dubin, A, Potempa, J, Holak, T.A. | | Deposit date: | 2005-08-23 | | Release date: | 2005-09-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Functional and Structural Characterization of Spl Proteases from Staphylococcus aureus
J.Mol.Biol., 358, 2006
|
|
2W7U
 
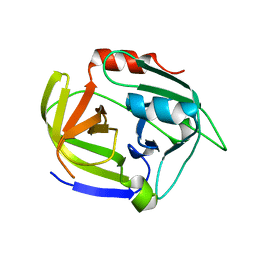 | | SplA serine protease of Staphylococcus aureus (2.4A) | | Descriptor: | SERINE PROTEASE SPLA | | Authors: | Stec-Niemczyka, J, Pustelny, K, Kisielewska, M, Bista, M, Boulware, K.T, Stennicke, H.R, Thogersen, I.B, Daugherty, P.S, Enghild, J.J, Popowicz, G.M, Dubin, A, Potempa, J, Dubin, G. | | Deposit date: | 2008-12-30 | | Release date: | 2010-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural and Functional Characterization of Spla, an Exclusively Specific Protease of Staphylococcus Aureus.
Biochem.J., 419, 2009
|
|
2W7S
 
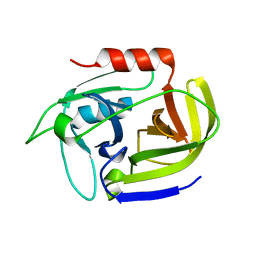 | | SplA serine protease of Staphylococcus aureus (1.8A) | | Descriptor: | SERINE PROTEASE SPLA | | Authors: | Stec-Niemczyka, J, Pustelny, K, Kisielewska, M, Bista, M, Boulware, K.T, Stennicke, H.R, Thogersen, I.B, Daugherty, P.S, Enghild, J.J, Popowicz, G.M, Dubin, A, Potempa, J, Dubin, G. | | Deposit date: | 2008-12-30 | | Release date: | 2010-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Functional Characterization of Spla, an Exclusively Specific Protease of Staphylococcus Aureus
Biochem.J., 419, 2009
|
|
4K1T
 
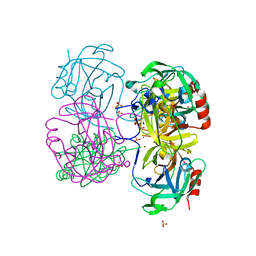 | | Gly-Ser-SplB protease from Staphylococcus aureus at 1.60 A resolution | | Descriptor: | CHLORIDE ION, SULFATE ION, Serine protease SplB, ... | | Authors: | Zdzalik, M, Pustelny, K, Stec-Niemczyk, J, Cichon, P, Czarna, A, Popowicz, G, Drag, M, Wladyka, B, Potempa, J, Dubin, A, Dubin, G. | | Deposit date: | 2013-04-05 | | Release date: | 2014-04-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Staphylococcal SplB Serine Protease Utilizes a Novel Molecular Mechanism of Activation.
J.Biol.Chem., 289, 2014
|
|
4K1S
 
 | | Gly-Ser-SplB protease from Staphylococcus aureus at 1.96 A resolution | | Descriptor: | Serine protease SplB | | Authors: | Zdzalik, M, Pustelny, K, Stec-Niemczyk, J, Cichon, P, Czarna, A, Popowicz, G, Drag, M, Wladyka, B, Potempa, J, Dubin, A, Dubin, G. | | Deposit date: | 2013-04-05 | | Release date: | 2014-04-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Staphylococcal SplB Serine Protease Utilizes a Novel Molecular Mechanism of Activation.
J.Biol.Chem., 289, 2014
|
|
8GH2
 
 | | Structure of Trypanosoma (MDH)4-(Pex5)2, close conformation | | Descriptor: | Peroxisome targeting signal 1 receptor, malate dehydrogenase | | Authors: | Sonani, R.R, Artur, B, Jemiola-Rzeminska, M, Lipinski, O, Patel, S.N, Sood, T, Dubin, G. | | Deposit date: | 2023-03-09 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Structure of Trypanosoma (MDH)4-(Pex5)2, close conformation
To Be Published
|
|
8GGH
 
 | | Structure of Trypanosoma (MDH)4-PEX5, distal conformation | | Descriptor: | Peroxisome targeting signal 1 receptor, malate dehydrogenase | | Authors: | Sonani, R.R, Artur, B, Jemiola-Rzeminska, M, Lipinski, O, Patel, S.N, Sood, T, Dubin, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structure of Trypanosoma (MDH)4-PEX5, distal conformation
To Be Published
|
|
8GH3
 
 | | Structure of Trypanosoma (MDH)4-(Pex5)2, distal conformation | | Descriptor: | Peroxisome targeting signal 1 receptor, malate dehydrogenase | | Authors: | Sonani, R.R, Artur, B, Jemiola-Rzeminska, M, Lipinski, O, Patel, S.N, Sood, T, Dubin, G. | | Deposit date: | 2023-03-09 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Structure of Trypanosoma (MDH)4-(Pex5)2, distal conformation
To Be Published
|
|
8GGD
 
 | | Structure of Trypanosoma (MDH)4-Pex5, close conformation | | Descriptor: | Peroxisome targeting signal 1 receptor, malate dehydrogenase | | Authors: | Sonani, R.R, Artur, B, Jemiola-Rzeminska, M, Lipinski, O, Patel, S.N, Sood, T, Dubin, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structure of Trypanosoma (MDH)4-Pex5, close conformation
To Be Published
|
|
8GI0
 
 | | Structure of Trypanosoma docking complex | | Descriptor: | Peroxisomal membrane protein PEX14, Peroxisome targeting signal 1 receptor, malate dehydrogenase | | Authors: | Sonani, R.R, Artur, B, Jemiola-Rzeminska, M, Lipinski, O, Patel, S.N, Sood, T, Dubin, G. | | Deposit date: | 2023-03-13 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of Trypanosoma docking complex
To Be Published
|
|
4INK
 
 | | Crystal structure of SplD protease from Staphylococcus aureus at 1.56 A resolution | | Descriptor: | Serine protease SplD | | Authors: | Zdzalik, M, Kalinska, M, Cichon, P, Wysocka, M, Stec-Niemczyk, J, Stennicke, H.R, Jabaiah, A, Markiewicz, M, Wladyka, B, Daugherty, P.S, Lesner, A, Rolka, K, Dubin, A, Potempa, J, Dubin, G. | | Deposit date: | 2013-01-04 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Biochemical and Structural Characterization of SplD Protease from Staphylococcus aureus.
Plos One, 8, 2013
|
|
4INL
 
 | | Crystal structure of SplD protease from Staphylococcus aureus at 2.1 A resolution | | Descriptor: | Serine protease SplD | | Authors: | Cichon, P, Zdzalik, M, Kalinska, M, Wysocka, M, Stec-Niemczyk, J, Stennicke, H.R, Jabaiah, A, Markiewicz, M, Wladyka, B, Daugherty, P.S, Lesner, A, Rolka, K, Dubin, A, Potempa, J, Dubin, G. | | Deposit date: | 2013-01-04 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical and Structural Characterization of SplD Protease from Staphylococcus aureus.
Plos One, 8, 2013
|
|
7Z35
 
 | |
7Q84
 
 | | Crystal structure of human peroxisomal acyl-Co-A oxidase 1a, apo-form | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Sonani, R.R, Blat, A, Dubin, G. | | Deposit date: | 2021-11-10 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of apo- and FAD-bound human peroxisomal acyl-CoA oxidase provide mechanistic basis explaining clinical observations.
Int.J.Biol.Macromol., 205, 2022
|
|
7Q86
 
 | |
7QOZ
 
 | |
7QRC
 
 | | X-ray structure of Trypanosoma cruzi PEX14 in complex with a PEX5-PEX14 PPI inhibitor | | Descriptor: | GLYCEROL, Peroxin-14, ~{N}-(5-ethyl-6-oxidanylidene-benzo[b][1,4]benzothiazepin-2-yl)-2-(4-fluorophenyl)ethanamide | | Authors: | Napolitano, V, Popowicz, G.M, Dawidowski, M, Dubin, G. | | Deposit date: | 2022-01-10 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure-based design, synthesis and evaluation of a novel family of PEX5-PEX14 interaction inhibitors against Trypanosoma.
Eur.J.Med.Chem., 243, 2022
|
|
7R2V
 
 | | Structure of nsp14 from SARS-CoV-2 in complex with SAH | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, Proofreading exoribonuclease nsp14, ... | | Authors: | Czarna, A, Plewka, J, Kresik, L, Matsuda, A, Abdulkarim, K, Robinson, C, OByrne, S, Cunningham, F, Georgiou, I, Pachota, M, Popowicz, G.M, Wyatt, P.G, Dubin, G, Pyrc, K. | | Deposit date: | 2022-02-06 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Refolding of lid subdomain of SARS-CoV-2 nsp14 upon nsp10 interaction releases exonuclease activity.
Structure, 30, 2022
|
|
7Z1G
 
 | | Crystal structure of nonphosphorylated (Tyr216) GSK3b in complex with CX-4945 | | Descriptor: | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, Glycogen synthase kinase-3 beta, IMIDAZOLE, ... | | Authors: | Grygier, P, Pustelny, K, Dubin, G, Czarna, A. | | Deposit date: | 2022-02-24 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of nonphosphorylated (Tyr216) GSK3b kinase in complex with CX-4945
To Be Published
|
|
