4ARY
 
 | | Lepidopteran-specific toxin Cry1Ac in complex with receptor specificity determinant GalNAc | | Descriptor: | 1,3-DIAMINOPROPANE, 2-acetamido-2-deoxy-beta-D-galactopyranose, PESTICIDAL CRYSTAL PROTEIN CRY1AC | | Authors: | Derbyshire, D.J, Carroll, J, Ellar, D.J, Li, J. | | Deposit date: | 2012-04-27 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural Basis of Galnac-Dependent Receptor Recognition by B. Thuringiensis Toxin Cry1Ac
To be Published
|
|
4ARX
 
 | | Lepidoptera-specific toxin Cry1Ac from Bacillus thuringiensis ssp. kurstaki HD-73 | | Descriptor: | 1,3-DIAMINOPROPANE, GLYCEROL, PESTICIDAL CRYSTAL PROTEIN CRY1AC | | Authors: | Derbyshire, D.J, Carroll, J, Ellar, D.J, Li, J. | | Deposit date: | 2012-04-27 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis of Galnac-Dependent Receptor Recognition by B. Thuringiensis Toxin Cry1Ac
To be Published
|
|
1GZH
 
 | |
6QLM
 
 | | Cathepsin-K in complex with MIV-701 | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, Cathepsin K, GLYCEROL, ... | | Authors: | Derbyshire, D.J. | | Deposit date: | 2019-02-01 | | Release date: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Successful development of 3-oxohexahydrofuropyrrole amino acid amides as inhibitors of Cathepsin-K.
To Be Published
|
|
6QM0
 
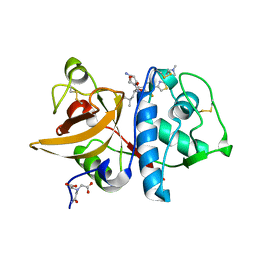 | | Cathepsin-K in complex with amino-oxaazabicyclo[3.3.0]octanyl containing inhibitor | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, Cathepsin K, ~{N}-[(2~{S})-1-[(3~{R},3~{a}~{R},6~{R},6~{a}~{R})-6-azanyl-3-oxidanyl-2,3,3~{a},5,6,6~{a}-hexahydrofuro[3,2-b]pyrrol-4-yl]-4-methyl-1-oxidanylidene-pentan-2-yl]-3-fluoranyl-4-[2-(4-methylpiperazin-1-yl)-1,3-thiazol-4-yl]benzamide | | Authors: | Derbyshire, D.J. | | Deposit date: | 2019-02-01 | | Release date: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Successful development of 3-oxohexahydrofuropyrrole amino acid amides as inhibitors of Cathepsin-K.
To Be Published
|
|
6QLX
 
 | | Cathepsin-K in complex with fluoro-oxa-azabicyclo[3.3.0]octanyl containing inhibitor | | Descriptor: | Cathepsin K, ~{N}-[(2~{S})-1-[(3~{R},3~{a}~{R},6~{S},6~{a}~{S})-6-fluoranyl-3-oxidanyl-2,3,3~{a},5,6,6~{a}-hexahydrofuro[3,2-b]pyrrol-4-yl]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-thiophen-2-yl-benzamide | | Authors: | Derbyshire, D.J. | | Deposit date: | 2019-02-01 | | Release date: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Successful development of 3-oxohexahydrofuropyrrole amino acid amides as inhibitors of Cathepsin-K.
To Be Published
|
|
6QLW
 
 | | Cathepsin-K in complex with MIV-710 | | Descriptor: | CHLORIDE ION, Cathepsin K, SODIUM ION, ... | | Authors: | Derbyshire, D.J. | | Deposit date: | 2019-02-01 | | Release date: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Successful development of 3-oxohexahydrofuropyrrole amino acid amides as inhibitors of Cathepsin-K.
To Be Published
|
|
6QL8
 
 | | Cathepsin-K in complex with MIV-711 | | Descriptor: | Cathepsin K, NITRATE ION, ~{N}-[(2~{S})-1-[(3~{R},3~{a}~{R},6~{R},6~{a}~{R})-6-ethynyl-3-oxidanyl-2,3,3~{a},5,6,6~{a}-hexahydrofuro[3,2-b]pyrrol-4-yl]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-[5-fluoranyl-2-(4-methylpiperazin-1-yl)-1,3-thiazol-4-yl]benzamide | | Authors: | Derbyshire, D.J. | | Deposit date: | 2019-01-31 | | Release date: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Successful development of 3-oxohexahydrofuropyrrole amino acid amides as inhibitors of Cathepsin-K.
To Be Published
|
|
3UVC
 
 | | MMP12 in a complex with the dimeric adduct: 5-(5-phenylhydantoin)-5-phenylhydantoin | | Descriptor: | (4R,4'S)-4,4'-diphenyl-4,4'-biimidazolidine-2,2',5,5'-tetrone, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Derbyshire, D.J, Danielson, H, Nystrum, S. | | Deposit date: | 2011-11-29 | | Release date: | 2013-01-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Characterization of fragments interacting with MMP-12
To be Published
|
|
2W0D
 
 | | Does a Fast Nuclear Magnetic Resonance Spectroscopy- and X-Ray Crystallography Hybrid Approach Provide Reliable Structural Information of Ligand-Protein Complexes? A Case Study of Metalloproteinases. | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Isaksson, J, Nystrom, S, Derbyshire, D.J, Wallberg, H, Agback, T, Kovacs, H, Bertini, I, Felli, I.C. | | Deposit date: | 2008-08-13 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Does a Fast Nuclear Magnetic Resonance Spectroscopy- and X-Ray Crystallography Hybrid Approach Provide Reliable Structural Information of Ligand-Protein Complexes? a Case Study of Metalloproteinases.
J.Med.Chem., 52, 2009
|
|
1H9O
 
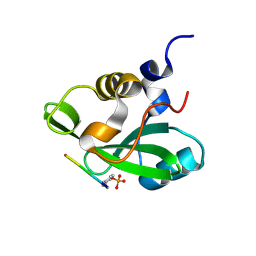 | | PHOSPHATIDYLINOSITOL 3-KINASE, P85-ALPHA SUBUNIT: C-TERMINAL SH2 DOMAIN COMPLEXED WITH A TYR751 PHOSPHOPEPTIDE FROM THE PDGF RECEPTOR, CRYSTAL STRUCTURE AT 1.79 A | | Descriptor: | BETA-PLATELET-DERIVED GROWTH FACTOR RECEPTOR, PHOSPHATIDYLINOSITOL 3-KINASE | | Authors: | Pauptit, R.A, Rowsell, S, Breeze, A.L, Murshudov, G.N, Dennis, C.A, Derbyshire, D.J, Weston, S.A. | | Deposit date: | 2001-03-14 | | Release date: | 2001-03-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | NMR Trial Models: Experiences with the Colicin Immunity Protein Im7 and the P85Alpha C-Terminal Sh2-Peptide Complex
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1IFT
 
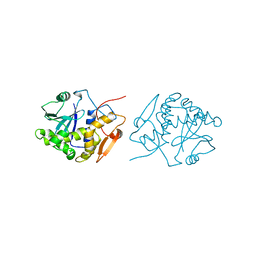 | | RICIN A-CHAIN (RECOMBINANT) | | Descriptor: | RICIN | | Authors: | Weston, S.A, Tucker, A.D, Thatcher, D.R, Derbyshire, D.J, Pauptit, R.A. | | Deposit date: | 1996-07-05 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray structure of recombinant ricin A-chain at 1.8 A resolution.
J.Mol.Biol., 244, 1994
|
|
1IFS
 
 | | RICIN A-CHAIN (RECOMBINANT) COMPLEX WITH ADENOSINE (ADENOSINE BECOMES ADENINE IN THE COMPLEX) | | Descriptor: | ADENINE, RICIN | | Authors: | Weston, S.A, Tucker, A.D, Thatcher, D.R, Derbyshire, D.J, Pauptit, R.A. | | Deposit date: | 1996-07-05 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of recombinant ricin A-chain at 1.8 A resolution.
J.Mol.Biol., 244, 1994
|
|
1IFU
 
 | | RICIN A-CHAIN (RECOMBINANT) COMPLEX WITH FORMYCIN | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, RICIN | | Authors: | Weston, S.A, Tucker, A.D, Thatcher, D.R, Derbyshire, D.J, Pauptit, R.A. | | Deposit date: | 1996-07-05 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structure of recombinant ricin A-chain at 1.8 A resolution.
J.Mol.Biol., 244, 1994
|
|
8AWI
 
 | | Crystal structure of Human Transthyretin at 1.15 Angstrom resolution | | Descriptor: | SODIUM ION, Transthyretin | | Authors: | Derbyshire, D.J, Hammarstrom, P, von Castelmur, E, Begum, A. | | Deposit date: | 2022-08-29 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Transthyretin Binding Mode Dichotomy of Fluorescent trans -Stilbene Ligands.
Acs Chem Neurosci, 14, 2023
|
|
7Q9N
 
 | | Transthyretin complexed with (E)-4-(2-(naphthalen-2-yl)vinyl)benzene-1,2-diol | | Descriptor: | 4-[(~{E})-2-naphthalen-2-ylethenyl]benzene-1,2-diol, Transthyretin | | Authors: | Derbyshire, D.J, Hammarstrom, P, von Castelmur, E, Begum, A. | | Deposit date: | 2021-11-12 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Transthyretin Binding Mode Dichotomy of Fluorescent trans -Stilbene Ligands.
Acs Chem Neurosci, 14, 2023
|
|
7Q9L
 
 | | Transthyretin complexed with (E)-4-(2-(naphthalen-1-yl)vinyl)benzene-1,2-diol | | Descriptor: | 4-[(~{E})-2-naphthalen-1-ylethenyl]benzene-1,2-diol, GLYCEROL, SODIUM ION, ... | | Authors: | Derbyshire, D.J, Hammarstrom, P, von Castelmur, E, Begum, A. | | Deposit date: | 2021-11-12 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Transthyretin Binding Mode Dichotomy of Fluorescent trans -Stilbene Ligands.
Acs Chem Neurosci, 14, 2023
|
|
7Q9O
 
 | | Complex of Transthyretin with resveratrol exhibits multiple binding modes | | Descriptor: | GLYCEROL, RESVERATROL, SODIUM ION, ... | | Authors: | Derbyshire, D.J, Hammarstrom, P, von Castelmur, E, Begum, A. | | Deposit date: | 2021-11-12 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Transthyretin Binding Mode Dichotomy of Fluorescent trans -Stilbene Ligands.
Acs Chem Neurosci, 14, 2023
|
|
1AJ6
 
 | |
1UQ5
 
 | | RICIN A-CHAIN (RECOMBINANT) N122A MUTANT | | Descriptor: | ACETATE ION, RICIN, SULFATE ION | | Authors: | Marsden, C.J, Fulop, V. | | Deposit date: | 2003-10-15 | | Release date: | 2004-01-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Effect of Mutations Surrounding and within the Active Site on the Catalytic Activity of Ricin a Chain
Eur.J.Biochem., 271, 2004
|
|
1UQ4
 
 | | RICIN A-CHAIN (RECOMBINANT) R213D MUTANT | | Descriptor: | RICIN, SULFATE ION | | Authors: | Marsden, C.J, Fulop, V. | | Deposit date: | 2003-10-15 | | Release date: | 2004-01-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Effect of Mutations Surrounding and within the Active Site on the Catalytic Activity of Ricin a Chain
Eur.J.Biochem., 271, 2004
|
|
1OBS
 
 | | STRUCTURE OF RICIN A CHAIN MUTANT | | Descriptor: | RICIN A CHAIN | | Authors: | Day, P.J, Ernst, S.R, Frankel, A.E, Monzingo, A.F, Pascal, J.M, Svinth, M, Robertus, J.D. | | Deposit date: | 1996-06-25 | | Release date: | 1997-06-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and activity of an active site substitution of ricin A chain.
Biochemistry, 35, 1996
|
|
1OBT
 
 | | STRUCTURE OF RICIN A CHAIN MUTANT, COMPLEX WITH AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, RICIN A CHAIN | | Authors: | Day, P.J, Ernst, S.R, Frankel, A.E, Monzingo, A.F, Pascal, J.M, Svinth, M, Robertus, J.D. | | Deposit date: | 1996-06-22 | | Release date: | 1997-06-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and activity of an active site substitution of ricin A chain.
Biochemistry, 35, 1996
|
|
