8TJT
 
 | | The Fab fragment of an anti-glucagon receptor (GCGR) antibody | | Descriptor: | anti-GCGR Fab heavy chain, anti-GCGR Fab light chain | | Authors: | Dai, J, Carter, P.J, Sudhamsu, J, Kung, J. | | Deposit date: | 2023-07-24 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Variable domain mutational analysis to probe the molecular mechanisms of high viscosity of an IgG 1 antibody.
Mabs, 16, 2024
|
|
6RMN
 
 | | DNA mismatch repair proteins MLH1 and MLH3 | | Descriptor: | DNA mismatch repair protein MLH1, DNA mismatch repair protein MLH3, ZINC ION | | Authors: | Dai, J, Chervy, P, Legrand, P, Ropars, V, Charbonnier, J.B. | | Deposit date: | 2019-05-07 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis of the dual role of the Mlh1-Mlh3 endonuclease in MMR and in meiotic crossover formation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6SHX
 
 | | DNA mismatch repair proteins MLH1 and MLH3 | | Descriptor: | DNA mismatch repair protein MLH1, DNA mismatch repair protein MLH3, ZINC ION | | Authors: | Dai, J, Chervy, P, Legrand, P, Ropars, V, Charbonnier, J.B. | | Deposit date: | 2019-08-08 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis of the dual role of the Mlh1-Mlh3 endonuclease in MMR and in meiotic crossover formation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6SNS
 
 | | DNA mismatch repair proteins MLH1 and MLH3 | | Descriptor: | DNA mismatch repair protein MLH1, DNA mismatch repair protein MLH3, ZINC ION | | Authors: | Dai, J, Chervy, P, Legrand, P, Ropars, V, Charbonnier, J.B. | | Deposit date: | 2019-08-27 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular basis of the dual role of the Mlh1-Mlh3 endonuclease in MMR and in meiotic crossover formation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6SNV
 
 | | DNA mismatch repair proteins MLH1 and MLH3 | | Descriptor: | DNA mismatch repair protein MLH1, DNA mismatch repair protein MLH3, ZINC ION | | Authors: | Dai, J, Chervy, P, Legrand, P, Ropars, V, Charbonnier, J.B. | | Deposit date: | 2019-08-27 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis of the dual role of the Mlh1-Mlh3 endonuclease in MMR and in meiotic crossover formation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8EAW
 
 | |
6X0Q
 
 | |
6X0R
 
 | |
9B4H
 
 | | Chlamydomonas reinhardtii mastigoneme filament | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C-type lectin domain-containing protein, Tyrosine-protein kinase ephrin type A/B receptor-like domain-containing protein, ... | | Authors: | Dai, J, Ma, M, Zhang, R, Brown, A. | | Deposit date: | 2024-03-20 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mastigoneme structure reveals insights into the O-linked glycosylation code of native hydroxyproline-rich helices.
Cell, 2024
|
|
1X95
 
 | | Solution structure of the DNA-hexamer ATGCAT complexed with DNA Bis-intercalating Anticancer Drug XR5944 (MLN944) | | Descriptor: | 1-METHYL-9-[12-(9-METHYLPHENAZIN-10-IUM-1-YL)-12-OXO-2,11-DIAZA-5,8-DIAZONIADODEC-1-ANOYL]PHENAZIN-10-IUM, 5'-D(*AP*TP*GP*CP*AP*T)-3' | | Authors: | Dai, J, Punchihewa, C, Mistry, P, Ooi, A.T, Yang, D. | | Deposit date: | 2004-08-19 | | Release date: | 2004-09-28 | | Last modified: | 2014-09-17 | | Method: | SOLUTION NMR | | Cite: | Novel DNA bis-intercalation by MLN944, a potent clinical bisphenazine anticancer drug.
J.Biol.Chem., 279, 2004
|
|
1S18
 
 | | Structure and protein design of human apyrase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Dai, J, Liu, J, Deng, Y, Smith, T.M, Lu, M. | | Deposit date: | 2004-01-05 | | Release date: | 2004-03-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and protein design of a human platelet function inhibitor.
Cell(Cambridge,Mass.), 116, 2004
|
|
1S1D
 
 | | Structure and protein design of human apyrase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Dai, J, Liu, J, Deng, Y, Smith, T.M, Lu, M. | | Deposit date: | 2004-01-06 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and protein design of a human platelet function inhibitor.
Cell(Cambridge,Mass.), 116, 2004
|
|
2F8U
 
 | | G-quadruplex structure formed in human Bcl-2 promoter, hybrid form | | Descriptor: | 5'-D(*GP*GP*GP*CP*GP*CP*GP*GP*GP*AP*GP*GP*AP*AP*TP*TP*GP*GP*GP*CP*GP*GP*G)-3' | | Authors: | Dai, J, Chen, D, Carver, M, Yang, D. | | Deposit date: | 2005-12-03 | | Release date: | 2006-11-07 | | Last modified: | 2014-09-17 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the major G-quadruplex structure formed in the human BCL2 promoter region.
Nucleic Acids Res., 34, 2006
|
|
2HY9
 
 | | Human telomere DNA quadruplex structure in K+ solution hybrid-1 form | | Descriptor: | DNA (26-MER) | | Authors: | Dai, J, Punchihewa, C, Jones, R.A, Hurley, L, Yang, D. | | Deposit date: | 2006-08-04 | | Release date: | 2007-04-10 | | Last modified: | 2014-09-17 | | Method: | SOLUTION NMR | | Cite: | Structure of the intramolecular human telomeric G-quadruplex in potassium solution: a novel adenine triple formation.
Nucleic Acids Res., 35, 2007
|
|
2JPZ
 
 | | Human telomere DNA quadruplex structure in K+ solution hybrid-2 form | | Descriptor: | DNA (26-MER) | | Authors: | Dai, J, Carver, M, Punchihewa, C, Jones, R, Yang, D. | | Deposit date: | 2007-05-25 | | Release date: | 2007-12-04 | | Last modified: | 2014-10-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the Hybrid-2 type intramolecular human telomeric G-quadruplex in K+ solution: insights into structure polymorphism of the human telomeric sequence
Nucleic Acids Res., 35, 2007
|
|
2L7V
 
 | | Quindoline/G-quadruplex complex | | Descriptor: | DNA (5'-D(*TP*GP*AP*GP*GP*GP*TP*GP*GP*GP*TP*AP*GP*GP*GP*TP*GP*GP*GP*TP*AP*A)-3'), N,N-diethyl-N'-(10H-indolo[3,2-b]quinolin-11-yl)ethane-1,2-diamine, POTASSIUM ION | | Authors: | Dai, J, Carver, M, Mathad, R, Yang, D. | | Deposit date: | 2010-12-23 | | Release date: | 2011-11-09 | | Last modified: | 2011-11-16 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a 2:1 Quindoline-c-MYC G-Quadruplex: Insights into G-Quadruplex-Interactive Small Molecule Drug Design.
J.Am.Chem.Soc., 133, 2011
|
|
7DHP
 
 | | Crystal structure of MazF from Deinococcus radiodurans | | Descriptor: | Endoribonuclease MazF, SULFATE ION | | Authors: | Zhao, Y, Dai, J. | | Deposit date: | 2020-11-17 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | MazEF Toxin-Antitoxin System-Mediated DNA Damage Stress Response in Deinococcus radiodurans.
Front Genet, 12, 2021
|
|
4QQV
 
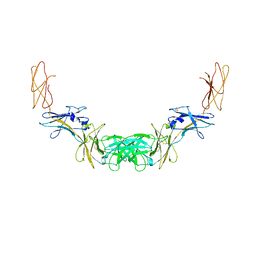 | | Extracellular domains of mouse IL-3 beta receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-3 receptor class 2 subunit beta | | Authors: | Jackson, C.J, Young, I.G, Murphy, J.M, Carr, P.D, Ewens, C.L, Dai, J, Ollis, D.L. | | Deposit date: | 2014-06-30 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Crystal structure of the mouse interleukin-3 beta-receptor: insights into interleukin-3 binding and receptor activation.
Biochem.J., 463, 2014
|
|
1JCC
 
 | | Crystal Structure of a Novel Alanine-Zipper Trimer at 1.7 A Resolution, V13A,L16A,V20A,L23A,V27A,M30A,V34A mutations | | Descriptor: | MAJOR OUTER MEMBRANE LIPOPROTEIN, ZINC ION | | Authors: | Liu, J, Dai, J, Lu, M. | | Deposit date: | 2001-06-08 | | Release date: | 2003-06-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Zinc-Mediated Helix Capping in A Triple-Helical Protein
Biochemistry, 42, 2003
|
|
1Z4O
 
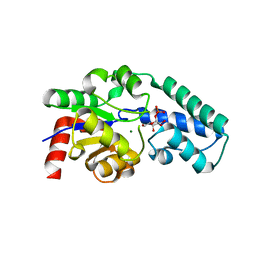 | | Structure of beta-phosphoglucomutase with inhibitor bound alpha-galactose 1-phosphate | | Descriptor: | 1-O-phosphono-alpha-D-galactopyranose, Beta-phosphoglucomutase, MAGNESIUM ION | | Authors: | Tremblay, L.W, Zhang, G, Dai, J, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2005-03-16 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Chemical Confirmation of a Pentavalent Phosphorane in Complex with beta-Phosphoglucomutase
J.Am.Chem.Soc., 127, 2005
|
|
1Z4N
 
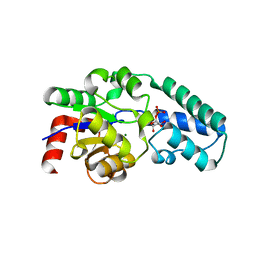 | | Structure of beta-phosphoglucomutase with inhibitor bound alpha-galactose 1-phosphate cocrystallized with Fluoride | | Descriptor: | 1-O-phosphono-alpha-D-galactopyranose, Beta-phosphoglucomutase, MAGNESIUM ION | | Authors: | Tremblay, L.W, Zhang, G, Dai, J, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2005-03-16 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Chemical Confirmation of a Pentavalent Phosphorane in Complex with beta-Phosphoglucomutase
J.Am.Chem.Soc., 127, 2005
|
|
1ZOL
 
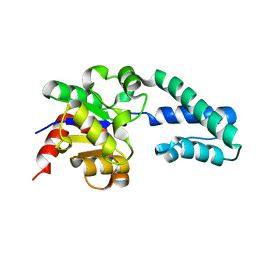 | | native beta-PGM | | Descriptor: | MAGNESIUM ION, beta-phosphoglucomutase | | Authors: | Zhang, G, Tremblay, L.W, Dai, J, Wang, L, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2005-05-13 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalytic cycling in beta-phosphoglucomutase: a kinetic and structural analysis
Biochemistry, 44, 2005
|
|
1SWV
 
 | | Crystal structure of the D12A mutant of phosphonoacetaldehyde hydrolase complexed with magnesium | | Descriptor: | MAGNESIUM ION, phosphonoacetaldehyde hydrolase | | Authors: | Zhang, G, Morais, M.C, Dai, J, Zhang, W, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2004-03-30 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Investigation of metal ion binding in phosphonoacetaldehyde hydrolase identifies sequence markers for metal-activated enzymes of the HAD enzyme superfamily
Biochemistry, 43, 2004
|
|
1SWW
 
 | | Crystal structure of the phosphonoacetaldehyde hydrolase D12A mutant complexed with magnesium and substrate phosphonoacetaldehyde | | Descriptor: | MAGNESIUM ION, PHOSPHONOACETALDEHYDE, phosphonoacetaldehyde hydrolase | | Authors: | Zhang, G, Morais, M.C, Dai, J, Zhang, W, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2004-03-30 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Investigation of metal ion binding in phosphonoacetaldehyde hydrolase identifies sequence markers for metal-activated enzymes of the HAD enzyme superfamily
Biochemistry, 43, 2004
|
|
1XAV
 
 | | Major G-quadruplex structure formed in human c-MYC promoter, a monomeric parallel-stranded quadruplex | | Descriptor: | 5'-D(*TP*GP*AP*GP*GP*GP*TP*GP*GP*GP*TP*AP*GP*GP*GP*TP*GP*GP*GP*TP*AP*A)-3', POTASSIUM ION | | Authors: | Ambrus, A, Chen, D, Dai, J, Jones, R.A, Yang, D. | | Deposit date: | 2004-08-26 | | Release date: | 2005-02-01 | | Last modified: | 2014-09-17 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the biologically relevant G-Quadruplex element in the human c-MYC promoter. Implications for G-quadruplex stabilization.
Biochemistry, 44, 2005
|
|
