5J02
 
 | | Structure of the lariat form of a chimeric derivative of the Oceanobacillus iheyensis group II intron in the presence of NH4+, MG2+ and an inactive 5' exon. | | Descriptor: | 5' EXON ANALOG (5'-R(*CP*UP*GP*UP*UP*AP*(5MU))-3'), AMMONIUM ION, GROUP II INTRON LARIAT, ... | | Authors: | Costa, M, Walbott, H, Monachello, D, Westhof, E, Michel, F. | | Deposit date: | 2016-03-26 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.493 Å) | | Cite: | Crystal structures of a group II intron lariat primed for reverse splicing.
Science, 354, 2016
|
|
5J01
 
 | | Structure of the lariat form of a chimeric derivative of the Oceanobacillus iheyensis group II intron in the presence of NH4+ and MG2+. | | Descriptor: | AMMONIUM ION, MAGNESIUM ION, group II intron lariat | | Authors: | Costa, M, Walbott, H, Monachello, D, Westhof, E, Michel, F. | | Deposit date: | 2016-03-26 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.39 Å) | | Cite: | Crystal structures of a group II intron lariat primed for reverse splicing.
Science, 354, 2016
|
|
1EA4
 
 | | TRANSCRIPTIONAL REPRESSOR COPG/22bp dsDNA COMPLEX | | Descriptor: | DNA (5'-D(*TP*AP*AP*CP*CP*GP*TP*GP *CP*AP*CP*TP*CP*AP*AP*TP*GP*CP*AP*AP*TP*C)-3'), DNA(5'-D(*AP*GP*AP*TP*TP*GP*CP*AP*TP *TP*GP*AP*GP*TP*GP*CP*AP*CP*GP*GP*TP*T)-3'), TRANSCRIPTIONAL REPRESSOR COPG | | Authors: | Gomis-Rueth, F.X, Costa, M, Sola, M, Acebo, P, Eritja, R, Espinosa, M, Solar, G.D, Coll, M. | | Deposit date: | 2000-11-05 | | Release date: | 2001-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Plasmid Transcriptional Repressor Copg Oligomerises to Render Helical Superstructures Unbound and in Complexes with Oligonucleotides
J.Mol.Biol., 310, 2001
|
|
1SUX
 
 | | CRYSTALLOGRAPHIC ANALYSIS OF THE COMPLEX BETWEEN TRIOSEPHOSPHATE ISOMERASE FROM TRYPANOSOMA CRUZI AND 3-(2-benzothiazolylthio)-1-propanesulfonic acid | | Descriptor: | 3-(2-BENZOTHIAZOLYLTHIO)-1-PROPANESULFONIC ACID, SULFATE ION, Triosephosphate isomerase, ... | | Authors: | Tellez-Valencia, A, Olivares-Illana, V, Hernandez-Santoyo, A, Perez-Montfort, R, Costas, M, Rodriguez-Romero, A, Tuena De Gomez-Puyou, M, Gomez-Puyou, A. | | Deposit date: | 2004-03-26 | | Release date: | 2004-08-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inactivation of triosephosphate isomerase from Trypanosoma cruzi by an agent that perturbs its dimer interface.
J.Mol.Biol., 341, 2004
|
|
1T3K
 
 | | NMR structure of a CDC25-like dual-specificity tyrosine phosphatase of Arabidopsis thaliana | | Descriptor: | Dual-specificity tyrosine phosphatase, ZINC ION | | Authors: | Landrieu, I, da Costa, M, De Veylder, L, Dewitte, F, Vandepoele, K, Hassan, S, Wieruszeski, J.M, Faure, J.D, Inze, D, Lippens, G. | | Deposit date: | 2004-04-27 | | Release date: | 2004-09-07 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | A small CDC25 dual-specificity tyrosine-phosphatase isoform in Arabidopsis thaliana.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
2JK2
 
 | | STRUCTURAL BASIS OF HUMAN TRIOSEPHOSPHATE ISOMERASE DEFICIENCY. CRYSTAL STRUCTURE OF THE WILD TYPE ENZYME. | | Descriptor: | TRIOSEPHOSPHATE ISOMERASE | | Authors: | Rodriguez-Almazan, C, Arreola-Alemon, R, Rodriguez-Larrea, D, Aguirre-Lopez, B, De Gomez-Puyou, M.T, Perez-Montfort, R, Costas, M, Gomez-Puyou, A, Torres-Larios, A. | | Deposit date: | 2008-06-22 | | Release date: | 2008-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Human Triosephosphate Isomerase Deficiency: Mutation E104D is Related to Alterations of a Conserved Water Network at the Dimer Interface.
J.Biol.Chem., 283, 2008
|
|
2VOM
 
 | | Structural basis of human triosephosphate isomerase deficiency. Mutation E104D and correlation to solvent perturbation. | | Descriptor: | TRIOSEPHOSPHATE ISOMERASE | | Authors: | Rodriguez-Almazan, C, Arreola-Alemon, R, Rodriguez-Larrea, D, Aguirre-Lopez, B, de Gomez-Puyou, M.T, Perez-Montfort, R, Costas, M, Gomez-Puyou, A, Torres-Larios, A. | | Deposit date: | 2008-02-19 | | Release date: | 2008-06-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis of Human Triosephosphate Isomerase Deficiency: Mutation E104D is Related to Alterations of a Conserved Water Network at the Dimer Interface.
J.Biol.Chem., 283, 2008
|
|
3O76
 
 | |
6NEE
 
 | |
2CJJ
 
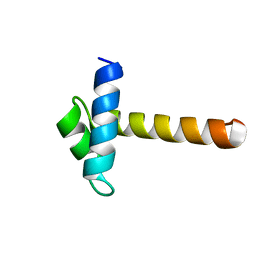 | | Crystal Structure of the MYB domain of the RAD transcription factor from Antirrhinum majus | | Descriptor: | RADIALIS | | Authors: | Stevenson, C.E.M, Burton, N, Costa, M.M, Nath, U, Dixon, R.A, Coen, E.S, Lawson, D.M. | | Deposit date: | 2006-04-04 | | Release date: | 2006-10-25 | | Last modified: | 2017-07-12 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Myb Domain of the Rad Transcription Factor from Antirrhinum Majus.
Proteins: Struct., Funct., Bioinf., 65, 2006
|
|
5V7U
 
 | |
5V7R
 
 | |
4Y9A
 
 | |
4Y8F
 
 | |
4Y96
 
 | | Crystal structure of Triosephosphate Isomerase from Gemmata obscuriglobus | | Descriptor: | CALCIUM ION, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Romero-Romero, S, Rodriguez-Romero, A, Fernandez-Velasco, D.A. | | Deposit date: | 2015-02-17 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | Reversibility and two state behaviour in the thermal unfolding of oligomeric TIM barrel proteins.
Phys Chem Chem Phys, 17, 2015
|
|
4Y90
 
 | | Crystal structure of Triosephosphate Isomerase from Deinococcus radiodurans | | Descriptor: | CALCIUM ION, GLYCEROL, SODIUM ION, ... | | Authors: | Romero-Romero, S, Rodriguez-Romero, A, Fernadez-Velasco, D.A. | | Deposit date: | 2015-02-16 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Reversibility and two state behaviour in the thermal unfolding of oligomeric TIM barrel proteins.
Phys Chem Chem Phys, 17, 2015
|
|
5CEH
 
 | | Structure of histone lysine demethylase KDM5A in complex with selective inhibitor | | Descriptor: | 7-oxo-5-phenyl-6-(propan-2-yl)-4,7-dihydropyrazolo[1,5-a]pyrimidine-3-carbonitrile, Lysine-specific demethylase 5A, NICKEL (II) ION, ... | | Authors: | Kiefer, J.R, Vinogradova, M. | | Deposit date: | 2015-07-06 | | Release date: | 2016-05-18 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | An inhibitor of KDM5 demethylases reduces survival of drug-tolerant cancer cells.
Nat.Chem.Biol., 12, 2016
|
|
7NB5
 
 | |
6G7Z
 
 | |
6D6Z
 
 | |
3CC0
 
 | |
3CBX
 
 | |
3CBY
 
 | |
3CBZ
 
 | |
2OMA
 
 | |
