3ZTN
 
 | | STRUCTURE OF INFLUENZA A NEUTRALIZING ANTIBODY SELECTED FROM CULTURES OF SINGLE HUMAN PLASMA CELLS IN COMPLEX WITH HUMAN H1 INFLUENZA HAEMAGGLUTININ. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FI6V3 ANTIBODY LIGHT CHAIN, ... | | Authors: | Hubbard, P.A, Ritchie, A.J, Corti, D, Voss, J.E, Gamblin, S.J, Codoni, G, Macagno, A, Jarrossay, D, Pinna, D, Minola, A, Vanzetta, F, Silacci, C, Fernandez-Rodriguez, B.M, Agatic, G, Giacchetto-Sasselli, I, Vachieri, S.G, Sallusto, F, Collins, P.J, Haire, L.F, Temperton, N, Langedijk, J.P.M, Skehel, J.J, Lanzavecchia, A. | | Deposit date: | 2011-07-12 | | Release date: | 2011-08-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | A Neutralizing Antibody Selected from Plasma Cells that Binds to Group 1 and Group 2 Influenza a Hemagglutinins.
Science, 333, 2011
|
|
7RNJ
 
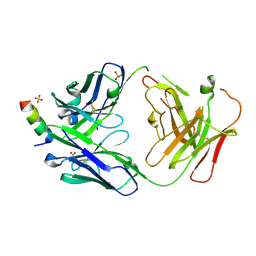 | | S2P6 Fab fragment bound to the SARS-CoV/SARS-CoV-2 spike stem helix peptide | | Descriptor: | Monoclonal antibody S2P6 Fab heavy chain, Monoclonal antibody S2P6 Fab light chain, SULFATE ION, ... | | Authors: | Snell, G, Czudnochowski, N, Croll, T.I, Nix, J.C, Corti, D, Cameroni, E, Pinto, D, Beltramello, M, Sauer, M.M, Veesler, D. | | Deposit date: | 2021-07-29 | | Release date: | 2021-08-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Broad betacoronavirus neutralization by a stem helix-specific human antibody.
Science, 373, 2021
|
|
6WPS
 
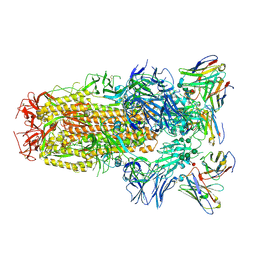 | | Structure of the SARS-CoV-2 spike glycoprotein in complex with the S309 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S309 neutralizing antibody heavy chain, ... | | Authors: | Pinto, D, Park, Y.J, Beltramello, M, Walls, A.C, Tortorici, M.A, Bianchi, S, Jaconi, S, Culap, K, Zatta, F, De Marco, A, Peter, A, Guarino, B, Spreafico, R, Cameroni, E, Case, J.B, Chen, R.E, Havenar-Daughton, C, Snell, G, Virgin, H.W, Lanzavecchia, A, Diamond, M.S, Fink, K, Veesler, D, Corti, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2020-04-27 | | Release date: | 2020-05-27 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody.
Nature, 583, 2020
|
|
6WS6
 
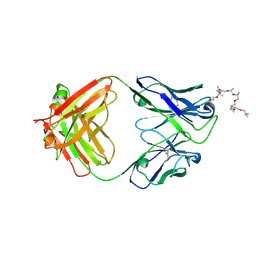 | | Structural and functional analysis of a potent sarbecovirus neutralizing antibody | | Descriptor: | O-(O-(2-AMINOPROPYL)-O'-(2-METHOXYETHYL)POLYPROPYLENE GLYCOL 500), S309 antigen-binding (Fab) fragment, heavy chain, ... | | Authors: | Pinto, D, Park, Y.J, Beltramello, M, Walls, A.C, Tortorici, M.A, Bianchi, S, Jaconi, S, Culap, K, Zatta, F, Marco, A.D, Peter, A, Guarino, B, Spreafico, R, Cameroni, E, Case, J.B, Chen, R.E, Havenar-Daughton, C, Snell, G, Telenti, A, Virgin, H.W, Lanzavecchia, A, Diamond, M.S, Fink, K, Veesler, D, Corti, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2020-04-30 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody.
Nature, 583, 2020
|
|
6WPT
 
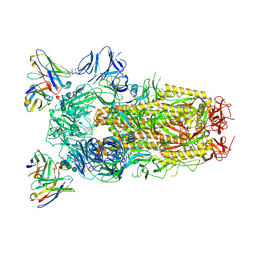 | | Structure of the SARS-CoV-2 spike glycoprotein in complex with the S309 neutralizing antibody Fab fragment (open state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S309 neutralizing antibody heavy chain, ... | | Authors: | Pinto, D, Park, Y.J, Beltramello, M, Walls, A.C, Tortorici, M.A, Bianchi, S, Jaconi, S, Culap, K, Zatta, F, De Marco, A, Peter, A, Guarino, B, Spreafico, R, Cameroni, E, Case, J.B, Chen, R.E, Havenar-Daughton, C, Snell, G, Virgin, H.W, Lanzavecchia, A, Diamond, M.S, Fink, K, Veesler, D, Corti, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2020-04-27 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody.
Nature, 583, 2020
|
|
6LNU
 
 | | Cryo-EM structure of immature Zika virus | | Descriptor: | Genome polyprotein | | Authors: | Tan, T.Y, Fibriansah, G, Kostyuchenko, V.A, Ng, T.S, Lim, X.X, Lim, X.N, Shi, J, Morais, M.C, Corti, D, Lok, S.M. | | Deposit date: | 2020-01-02 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Capsid protein structure in Zika virus reveals the flavivirus assembly process.
Nat Commun, 11, 2020
|
|
6X79
 
 | | Prefusion SARS-CoV-2 S ectodomain trimer covalently stabilized in the closed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | McCallum, M, Walls, A.C, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2020-05-29 | | Release date: | 2020-08-19 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure-guided covalent stabilization of coronavirus spike glycoprotein trimers in the closed conformation.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7R6X
 
 | | SARS-CoV-2 spike receptor-binding domain (RBD) in complex with S2E12 Fab, S309 Fab, and S304 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Monoclonal antibody S2E12 Fab heavy chain, ... | | Authors: | Snell, G, Czudnochowski, N, Croll, T.I, Nix, J.C, Corti, D, Cameroni, E, Pinto, D, Beltramello, M. | | Deposit date: | 2021-06-23 | | Release date: | 2021-07-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape.
Nature, 597, 2021
|
|
2XRA
 
 | | crystal structure of the HK20 Fab in complex with a gp41 mimetic 5- Helix | | Descriptor: | HK20, HUMAN MONOCLONAL ANTIBODY HEAVY CHAIN, HUMAN MONOCLONAL ANTIBODY LIGHT CHAIN, ... | | Authors: | Sabin, C, Corti, D, Buzon, V, Seaman, M.S, Lutje Hulsik, D, Hinz, A, Vanzetta, F, Agatic, G, Silacci, C, Langedijk, J.P.M, Mainetti, L, Scarlatti, G, Sallusto, F, Weiss, R, Lanzavecchia, A, Weissenhorn, W. | | Deposit date: | 2010-09-13 | | Release date: | 2010-12-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and size-dependent neutralization properties of HK20, a human monoclonal antibody binding to the highly conserved heptad repeat 1 of gp41.
PLoS Pathog., 6, 2010
|
|
9ATM
 
 | | SARS-CoV-2 EG.5 RBD bound to the VIR-7229 and the S2H97 Fab fragments | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rietz, T, Park, Y.J, Errico, J, Czudnochowski, N, Nix, J.C, Corti, D, Snell, G, Marco, A.D, Pinto, D, Cameroni, E, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2024-02-27 | | Release date: | 2024-10-16 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A potent pan-sarbecovirus neutralizing antibody resilient to epitope diversification.
Cell, 187, 2024
|
|
9AU1
 
 | | SARS-CoV-2 XBB.1.5 RBD bound to the VIR-7229 and the S309 Fab fragments | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Rietz, T, Park, Y.J, Errico, J, Czudnochowski, N, Nix, J.C, Corti, D, Snell, G, Marco, A.D, Pinto, D, Cameroni, E, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-02-27 | | Release date: | 2024-10-16 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | A potent pan-sarbecovirus neutralizing antibody resilient to epitope diversification.
Cell, 187, 2024
|
|
6SNE
 
 | | crystal structure of LN01 Fab in complex with an HIV-1 gp41 peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, LN01 heavy chain, ... | | Authors: | Caillat, C, Pinto, D, Corti, D, Fenwick, C, Pantaleo, G, Weissenhorn, W. | | Deposit date: | 2019-08-23 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural Basis for Broad HIV-1 Neutralization by the MPER-Specific Human Broadly Neutralizing Antibody LN01.
Cell Host Microbe, 26, 2019
|
|
5A3I
 
 | | Crystal Structure of a Complex formed between FLD194 Fab and Transmissible Mutant H5 Haemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ANTI-HAEMAGGLUTININ HA1 FAB HEAVY CHAIN, ... | | Authors: | Xiong, X, Corti, D, Liu, J, Pinna, D, Foglierini, M, Calder, L.J, Martin, S.R, Lin, Y.P, Walker, P.A, Collins, P.J, Monne, I, Suguitan Jr, A.L, Santos, C, Temperton, N.J, Subbarao, K, Lanzavecchia, A, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2015-06-01 | | Release date: | 2015-07-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structures of Complexes Formed by H5 Influenza Hemagglutinin with a Potent Broadly Neutralizing Human Monoclonal Antibody.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6SND
 
 | | crystal structure of LN01 Fab in complex with an HIV-1 gp41 peptide | | Descriptor: | 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Caillat, C, Pinto, D, Corti, D, Fenwick, C, Pantaleo, G, Weissenhorn, W. | | Deposit date: | 2019-08-23 | | Release date: | 2019-11-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis for Broad HIV-1 Neutralization by the MPER-Specific Human Broadly Neutralizing Antibody LN01.
Cell Host Microbe, 26, 2019
|
|
6SNC
 
 | | crystal structure of LN01 Fab in complex with an HIV-1 gp41 peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, LNO1 Heavy Chain, ... | | Authors: | Caillat, C, Pinto, D, Corti, D, Fenwick, C, Pantaleo, G, Weissenhorn, W. | | Deposit date: | 2019-08-23 | | Release date: | 2019-11-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis for Broad HIV-1 Neutralization by the MPER-Specific Human Broadly Neutralizing Antibody LN01.
Cell Host Microbe, 26, 2019
|
|
8S6M
 
 | | SARS-CoV-2 BQ.1.1 RBD bound to the S2V29 and the S2H97 Fab fragments | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Errico, J.M, Park, Y.J, Rietz, T, Czudnochowski, N, Nix, J.C, Cameroni, E, Corti, D, Snell, G, Marco, A.D, Pinto, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2024-02-28 | | Release date: | 2024-10-23 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | A potent pan-sarbecovirus neutralizing antibody resilient to epitope diversification.
Cell, 187, 2024
|
|
7M7W
 
 | | Antibodies to the SARS-CoV-2 receptor-binding domain that maximize breadth and resistance to viral escape | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Monoclonal antibody S2H97 Fab heavy chain, Monoclonal antibody S2H97 Fab light chain, ... | | Authors: | Snell, G, Czudnochowski, N, Croll, T.I, Nix, J.C, Corti, D, Cameroni, E, Pinto, D, Beltramello, M. | | Deposit date: | 2021-03-29 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape.
Nature, 597, 2021
|
|
7R6W
 
 | | SARS-CoV-2 spike receptor-binding domain (RBD) in complex with S2X35 Fab and S309 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Snell, G, Czudnochowski, N, Hernandez, P, Nix, J.C, Croll, T.I, Corti, D, Cameroni, E, Pinto, D, Beltramello, M. | | Deposit date: | 2021-06-23 | | Release date: | 2021-07-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape.
Nature, 597, 2021
|
|
7U9G
 
 | | Rabies virus glycoprotein pre-fusion trimer in complex with neutralizing antibody RVA122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein, RVA122 Fab Heavy Chain, ... | | Authors: | Callaway, H.M, Zyla, D, Larrous, F, Dias de Melo, G, Hastie, K.M, Avalos, R.D, Agarwal, A, Bouhry, H, Corti, D, Saphire, E.O. | | Deposit date: | 2022-03-10 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structure of the rabies virus glycoprotein trimer bound to a prefusion-specific neutralizing antibody.
Sci Adv, 8, 2022
|
|
6NB3
 
 | | MERS-CoV complex with human neutralizing LCA60 antibody Fab fragment (state 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LCA60 heavy chain, ... | | Authors: | Walls, A.C, Xiong, X, Park, Y.J, Tortorici, M.A, Snijder, S, Quispe, J, Cameroni, E, Gopal, R, Mian, D, Lanzavecchia, A, Zambon, M, Rey, F.A, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion.
Cell, 176, 2019
|
|
6NB4
 
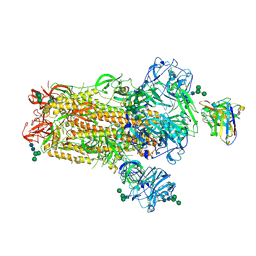 | | MERS-CoV S complex with human neutralizing LCA60 antibody Fab fragment (state 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LCA60 heavy chain, ... | | Authors: | Walls, A.C, Xiong, X, Park, Y.J, Tortorici, M.A, Snijder, S, Quispe, J, Cameroni, E, Gopal, R, Mian, D, Lanzavecchia, A, Zambon, M, Rey, F.A, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion.
Cell, 176, 2019
|
|
6NB7
 
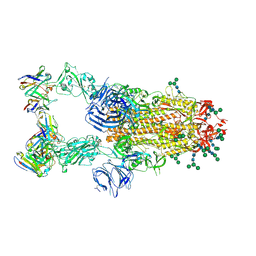 | | SARS-CoV complex with human neutralizing S230 antibody Fab fragment (state 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S230 heavy chain, ... | | Authors: | Walls, A.C, Xiong, X, Park, Y.J, Tortorici, M.A, Snijder, S, Quispe, J, Cameroni, E, Gopal, R, Mian, D, Lanzavecchia, A, Zambon, M, Rey, F.A, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-06 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion.
Cell, 176, 2019
|
|
6NB6
 
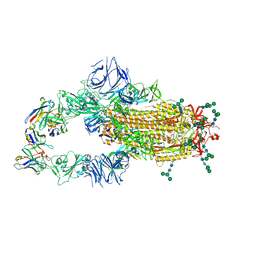 | | SARS-CoV complex with human neutralizing S230 antibody Fab fragment (state 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S230 heavy chain, ... | | Authors: | Walls, A.C, Xiong, X, Park, Y.J, Tortorici, M.A, Snijder, S, Quispe, J, Cameroni, E, Gopal, R, Mian, D, Lanzavecchia, A, Zambon, M, Rey, F.A, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-06 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion.
Cell, 176, 2019
|
|
6NB5
 
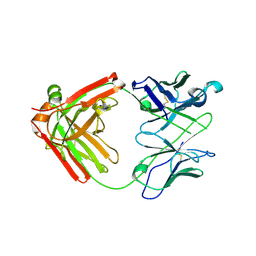 | | Crystal structure of anti- MERS-CoV human neutralizing LCA60 antibody Fab fragment | | Descriptor: | LCA60 antigen-binding (Fab) fragment, heavy chain, light chain | | Authors: | Walls, A.J, Xiong, X, Park, Y.J, Tortorici, M.A, Snijder, J, Quispe, J, Cameroni, E, Gopal, R, Dai, M, Lanzavecchia, A, Zambon, M, Rey, F.A, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion.
Cell, 176, 2019
|
|
6NB8
 
 | | Crystal structure of anti- SARS-CoV human neutralizing S230 antibody Fab fragment | | Descriptor: | S230 antigen-binding (Fab) fragment, heavy chain, light chain | | Authors: | Walls, A.J, Xiong, X, Park, Y.J, Tortorici, M.A, Snijder, J, Quispe, J, Cameroni, E, Gopal, R, Dai, M, Lanzavecchia, A, Zambon, M, Rey, F.A, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion.
Cell, 176, 2019
|
|
