1HVR
 
 | | RATIONAL DESIGN OF POTENT, BIOAVAILABLE, NONPEPTIDE CYCLIC UREAS AS HIV PROTEASE INHIBITORS | | Descriptor: | HIV-1 PROTEASE, [4R-(4ALPHA,5ALPHA,6BETA,7BETA)]-HEXAHYDRO-5,6-DIHYDROXY-1,3-BIS[2-NAPHTHYL-METHYL]-4,7-BIS(PHENYLMETHYL)-2H-1,3-DIAZEPIN-2-ONE | | Authors: | Chang, C.-H. | | Deposit date: | 1994-02-14 | | Release date: | 1995-01-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rational design of potent, bioavailable, nonpeptide cyclic ureas as HIV protease inhibitors.
Science, 263, 1994
|
|
2RCR
 
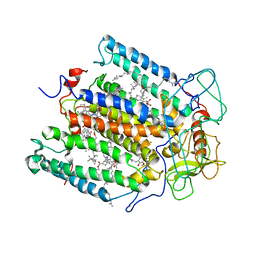 | | STRUCTURE OF THE MEMBRANE-BOUND PROTEIN PHOTOSYNTHETIC REACTION CENTER FROM RHODOBACTER SPHAEROIDES | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, Coenzyme Q10, ... | | Authors: | Chang, C.-H, Norris, J, Schiffer, M. | | Deposit date: | 1991-02-04 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the membrane-bound protein photosynthetic reaction center from Rhodobacter sphaeroides.
Biochemistry, 30, 1991
|
|
2P93
 
 | |
2P94
 
 | |
2P95
 
 | |
2D2W
 
 | |
3CEN
 
 | |
1LVE
 
 | |
2A3S
 
 | |
2C6Y
 
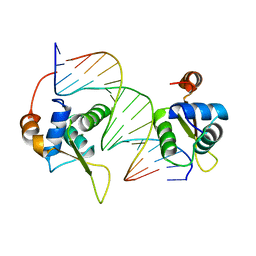 | | Crystal structure of interleukin enhancer-binding factor 1 bound to DNA | | Descriptor: | FORKHEAD BOX PROTEIN K2, INTERLEUKIN 2 PROMOTOR, MAGNESIUM ION | | Authors: | Tsai, K.-L, Huang, C.-Y, Chang, C.-H, Sun, Y.-J, Chuang, W.-J, Hsiao, C.-D. | | Deposit date: | 2005-11-15 | | Release date: | 2006-04-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Human Foxk1A-DNA Complex and its Implications on the Diverse Binding Specificity of Winged Helix/Forkhead Proteins.
J.Biol.Chem., 281, 2006
|
|
1DMP
 
 | | STRUCTURE OF HIV-1 PROTEASE COMPLEX | | Descriptor: | HIV-1 PROTEASE, [4-R-(-4-ALPHA,5-ALPHA,6-BETA,7-BETA)]-HEXAHYDRO-5,6-BIS(HYDROXY)-1,3-BIS([(3-AMINO)PHENYL]METHYL)-4,7-BIS(PHENYLMETHYL)-2H-1,3-DIAZEPINONE | | Authors: | Chang, C.-H. | | Deposit date: | 1996-11-01 | | Release date: | 1997-11-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Improved cyclic urea inhibitors of the HIV-1 protease: synthesis, potency, resistance profile, human pharmacokinetics and X-ray crystal structure of DMP 450.
Chem.Biol., 3, 1996
|
|
1HVH
 
 | | NONPEPTIDE CYCLIC CYANOGUANIDINES AS HIV PROTEASE INHIBITORS | | Descriptor: | HIV-1 PROTEASE, {[4-R(-4-ALPHA,5-ALPHA,6-BETA,7-BETA)]-HEXAHYDRO-5,6-BIS(HYDROXY)-1,3-BIS(4-HYDROXYMETHYL)METHYL]-4,7-BIS(PHENYLMETHYL) -2H-1,3-DIAZEPIN-2-YLIDENE]CYANAMIDE} | | Authors: | Chang, C.-H. | | Deposit date: | 1997-12-13 | | Release date: | 1998-12-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Nonpeptide cyclic cyanoguanidines as HIV-1 protease inhibitors: synthesis, structure-activity relationships, and X-ray crystal structure studies.
J.Med.Chem., 41, 1998
|
|
1HWR
 
 | | MOLECULAR RECOGNITION OF CYCLIC UREA HIV PROTEASE INHIBITORS | | Descriptor: | HIV-1 PROTEASE, [4-R-(4-ALPHA,6-BETA,7-BETA]-HEXAHYDRO-5,6-DI(HYDROXY)-1,3-DI(ALLYL)-4,7-BISPHENYLMETHYL)-2H-1,3-DIAZEPINONE | | Authors: | Chang, C.-H. | | Deposit date: | 1998-03-20 | | Release date: | 1999-03-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular recognition of cyclic urea HIV-1 protease inhibitors.
J.Biol.Chem., 273, 1998
|
|
1KIG
 
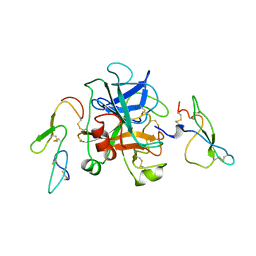 | | BOVINE FACTOR XA | | Descriptor: | ANTICOAGULANT PEPTIDE, FACTOR XA | | Authors: | Wei, A, Alexander, R, Chang, C.-H. | | Deposit date: | 1997-04-24 | | Release date: | 1998-10-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Unexpected binding mode of tick anticoagulant peptide complexed to bovine factor Xa.
J.Mol.Biol., 283, 1998
|
|
5LVE
 
 | |
3GIV
 
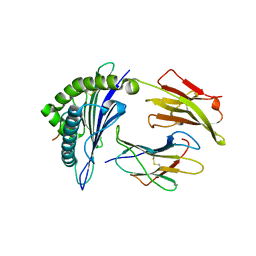 | | Antigen processing influences HIV-specific cytotoxic T lymphocyte immunodominance | | Descriptor: | Beta-2-microglobulin, HIV-1 peptide, HLA class I histocompatibility antigen, ... | | Authors: | Stewart-Jones, G, Iversen, A.K.N, Jones, E.Y. | | Deposit date: | 2009-03-06 | | Release date: | 2009-06-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Antigen processing influences HIV-specific cytotoxic T lymphocyte immunodominance
Nat.Immunol., 10, 2009
|
|
3MRU
 
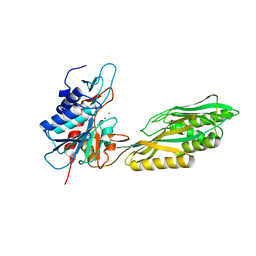 | | Crystal Structure of Aminoacylhistidine Dipeptidase from Vibrio alginolyticus | | Descriptor: | Aminoacyl-histidine dipeptidase, ZINC ION | | Authors: | Chang, C.-Y, Hsieh, Y.-C, Wu, T.-K, Chen, C.-J. | | Deposit date: | 2010-04-29 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure and mutational analysis of aminoacylhistidine dipeptidase from vibrio alginolyticus reveal a new architecture of M20 metallopeptidases
J.Biol.Chem., 285, 2010
|
|
4BJL
 
 | |
1MER
 
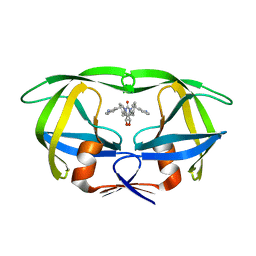 | | HIV-1 MUTANT (I84V) PROTEASE COMPLEXED WITH DMP450 | | Descriptor: | HIV-1 PROTEASE, [4-R-(-4-ALPHA,5-ALPHA,6-BETA,7-BETA)]-HEXAHYDRO-5,6-BIS(HYDROXY)-1,3-BIS([(3-AMINO)PHENYL]METHYL)-4,7-BIS(PHENYLMETHYL)-2H-1,3-DIAZEPINONE | | Authors: | Ala, P, Chang, C.-H. | | Deposit date: | 1997-04-11 | | Release date: | 1998-04-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis of HIV-1 protease drug resistance: structural analysis of mutant proteases complexed with cyclic urea inhibitors.
Biochemistry, 36, 1997
|
|
1MEU
 
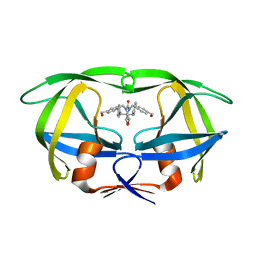 | | HIV-1 MUTANT (V82F, I84V) PROTEASE COMPLEXED WITH DMP323 | | Descriptor: | HIV-1 PROTEASE, [4-R-(-4-ALPHA,5-ALPHA,6-BETA,7-BETA)]-HEXAHYDRO-5,6-BIS(HYDROXY)-[1,3-BIS([4-HYDROXYMETHYL-PHENYL]METHYL)-4,7-BIS(PHEN YLMETHYL)]-2H-1,3-DIAZEPINONE | | Authors: | Ala, P, Chang, C.-H. | | Deposit date: | 1997-04-11 | | Release date: | 1998-04-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis of HIV-1 protease drug resistance: structural analysis of mutant proteases complexed with cyclic urea inhibitors.
Biochemistry, 36, 1997
|
|
1MET
 
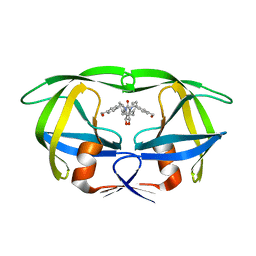 | | HIV-1 MUTANT (V82F) PROTEASE COMPLEXED WITH DMP323 | | Descriptor: | HIV-1 PROTEASE, [4-R-(-4-ALPHA,5-ALPHA,6-BETA,7-BETA)]-HEXAHYDRO-5,6-BIS(HYDROXY)-[1,3-BIS([4-HYDROXYMETHYL-PHENYL]METHYL)-4,7-BIS(PHEN YLMETHYL)]-2H-1,3-DIAZEPINONE | | Authors: | Ala, P, Chang, C.-H. | | Deposit date: | 1997-04-11 | | Release date: | 1998-04-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis of HIV-1 protease drug resistance: structural analysis of mutant proteases complexed with cyclic urea inhibitors.
Biochemistry, 36, 1997
|
|
1MES
 
 | | HIV-1 MUTANT (I84V) PROTEASE COMPLEXED WITH DMP323 | | Descriptor: | HIV-1 PROTEASE, [4-R-(-4-ALPHA,5-ALPHA,6-BETA,7-BETA)]-HEXAHYDRO-5,6-BIS(HYDROXY)-[1,3-BIS([4-HYDROXYMETHYL-PHENYL]METHYL)-4,7-BIS(PHEN YLMETHYL)]-2H-1,3-DIAZEPINONE | | Authors: | Ala, P, Chang, C.-H. | | Deposit date: | 1997-04-11 | | Release date: | 1998-04-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis of HIV-1 protease drug resistance: structural analysis of mutant proteases complexed with cyclic urea inhibitors.
Biochemistry, 36, 1997
|
|
1BJM
 
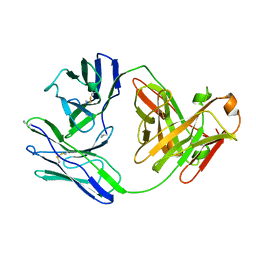 | |
1QBS
 
 | | HIV-1 PROTEASE INHIBITORS WIIH LOW NANOMOLAR POTENCY | | Descriptor: | HIV-1 PROTEASE, [4-R-(-4-ALPHA,5-ALPHA,6-BETA,7-BETA)]-HEXAHYDRO-5,6-BIS(HYDROXY)-[1,3-BIS([4-HYDROXYMETHYL-PHENYL]METHYL)-4,7-BIS(PHEN YLMETHYL)]-2H-1,3-DIAZEPINONE | | Authors: | Ala, P, Chang, C.-H. | | Deposit date: | 1997-04-25 | | Release date: | 1997-10-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cyclic HIV protease inhibitors: synthesis, conformational analysis, P2/P2' structure-activity relationship, and molecular recognition of cyclic ureas.
J.Med.Chem., 39, 1996
|
|
1QBR
 
 | | HIV-1 PROTEASE INHIBITORS WIIH LOW NANOMOLAR POTENCY | | Descriptor: | HIV-1 PROTEASE, [4R-(4ALPHA,5ALPHA,6BETA,7BETA)]-3,3'-[[TETRAHYDRO-5,6-DIHYDROXY-2-OXO-4,7-BIS(PHENYLMETHYL)-1H-1,3-DIAZEPINE-1,3(2H)-D IYL] BIS(METHYLENE)]BIS[N-2-THIAZOLYLBENZAMIDE] | | Authors: | Ala, P, Chang, C.-H. | | Deposit date: | 1997-04-25 | | Release date: | 1997-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cyclic urea amides: HIV-1 protease inhibitors with low nanomolar potency against both wild type and protease inhibitor resistant mutants of HIV.
J.Med.Chem., 40, 1997
|
|
