5A31
 
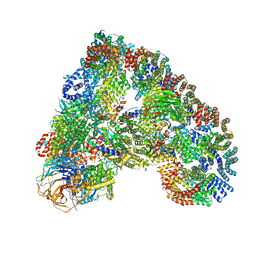 | | Structure of the human APC-Cdh1-Hsl1-UbcH10 complex. | | Descriptor: | ANAPHASE-PROMOTING COMPLEX SUBUNIT 1, ANAPHASE-PROMOTING COMPLEX SUBUNIT 10, ANAPHASE-PROMOTING COMPLEX SUBUNIT 11, ... | | Authors: | Chang, L, Zhang, Z, Yang, J, Mclaughlin, S.H, Barford, D. | | Deposit date: | 2015-05-26 | | Release date: | 2015-11-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Atomic Structure of the Apc/C and its Mechanism of Protein Ubiquitination.
Nature, 522, 2015
|
|
4UI9
 
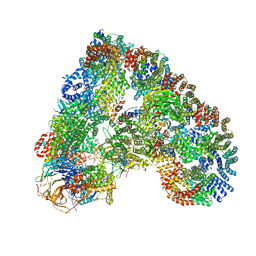 | | Atomic structure of the human Anaphase-Promoting Complex | | Descriptor: | ANAPHASE-PROMOTING COMPLEX SUBUNIT 1, ANAPHASE-PROMOTING COMPLEX SUBUNIT 10, ANAPHASE-PROMOTING COMPLEX SUBUNIT 11, ... | | Authors: | Chang, L, Zhang, Z, Yang, J, McLaughlin, S.H, Barford, D. | | Deposit date: | 2015-03-27 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Atomic Structure of the Apc and its Mechanism of Protein Ubiquitination
Nature, 522, 2015
|
|
6TGB
 
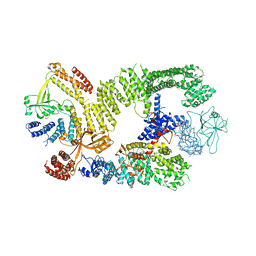 | | CryoEM structure of the binary DOCK2-ELMO1 complex | | Descriptor: | Dedicator of cytokinesis protein 2, Engulfment and cell motility protein 1 | | Authors: | Chang, L, Yang, J, Chang, J.H, Zhang, Z, Boland, A, McLaughlin, S.H, Abu-Thuraia, A, Killoran, R.C, Smith, M.J, Cote, J.F, Barford, D. | | Deposit date: | 2019-11-15 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structure of the DOCK2-ELMO1 complex provides insights into regulation of the auto-inhibited state.
Nat Commun, 11, 2020
|
|
6TGC
 
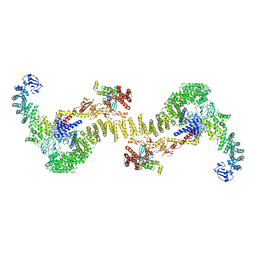 | | CryoEM structure of the ternary DOCK2-ELMO1-RAC1 complex. | | Descriptor: | Dedicator of cytokinesis protein 2, Engulfment and cell motility protein 1, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Chang, L, Yang, J, Chang, J.H, Zhang, Z, Boland, A, McLaughlin, S.H, Abu-Thuraia, A, Killoran, R.C, Smith, M.J, Cote, J.F, Barford, D. | | Deposit date: | 2019-11-15 | | Release date: | 2020-07-29 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of the DOCK2-ELMO1 complex provides insights into regulation of the auto-inhibited state.
Nat Commun, 11, 2020
|
|
1HQ3
 
 | | CRYSTAL STRUCTURE OF THE HISTONE-CORE-OCTAMER IN KCL/PHOSPHATE | | Descriptor: | CHLORIDE ION, HISTONE H2A-IV, HISTONE H2B, ... | | Authors: | Chantalat, L, Nicholson, J.M, Lambert, S.J, Reid, A.J, Donovan, M.J, Reynolds, C.D, Wood, C.M, Baldwin, J.P. | | Deposit date: | 2000-12-14 | | Release date: | 2001-01-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of the histone-core octamer in KCl/phosphate crystals at 2.15 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1QF8
 
 | | TRUNCATED FORM OF CASEIN KINASE II BETA SUBUNIT (2-182) FROM HOMO SAPIENS | | Descriptor: | CASEIN KINASE II, MAGNESIUM ION, ZINC ION | | Authors: | Chantalat, L, Leroy, D, Filhol, O, Nueda, A, Benitez, M.J, Chambaz, E, Cochet, C, Dideberg, O. | | Deposit date: | 1999-04-07 | | Release date: | 2000-04-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of the human protein kinase CK2 regulatory subunit reveals its zinc finger-mediated dimerization.
EMBO J., 18, 1999
|
|
1DXK
 
 | |
1E31
 
 | | SURVIVIN DIMER H. SAPIENS | | Descriptor: | APOPTOSIS INHIBITOR SURVIVIN, COBALT (II) ION, ZINC ION | | Authors: | Chantalat, L, Skoufias, D.A, Margolis, R.L, Dideberg, O. | | Deposit date: | 2000-06-04 | | Release date: | 2001-01-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal Structure of Human Survivin Reveals a Bow Tie-Shaped Dimer with Two Unusual Alpha-Helical Extensions
Mol.Cell, 6, 2000
|
|
1HGU
 
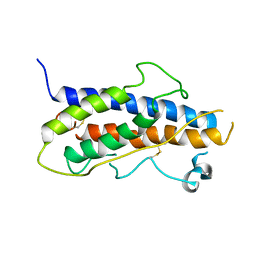 | | HUMAN GROWTH HORMONE | | Descriptor: | HUMAN GROWTH HORMONE | | Authors: | Chantalat, L, Jones, N, Korber, F, Navaza, J, Pavlovsky, A.G. | | Deposit date: | 1995-05-11 | | Release date: | 1995-12-07 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | THE CRYSTAL-STRUCTURE OF WILD-TYPE GROWTH-HORMONE AT 2.5 ANGSTROM RESOLUTION.
Protein Pept.Lett., 2, 1995
|
|
7JZY
 
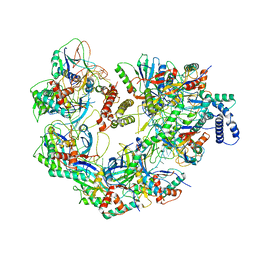 | | CryoEM structure of a CRISPR-Cas complex | | Descriptor: | AcrF9, CRISPR type I-F/YPEST-associated protein Csy3, CRISPR-associated protein Csy1, ... | | Authors: | Chang, L, Li, Z, Gabel, C. | | Deposit date: | 2020-09-02 | | Release date: | 2021-09-22 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | CryoEM structure of a CRISPR-Cas complex
To Be Published
|
|
6FX0
 
 | |
6NMA
 
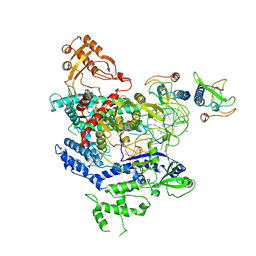 | | CryoEM structure of the LbCas12a-crRNA-AcrVA4 complex | | Descriptor: | AcrVA1, Cpf1, MAGNESIUM ION, ... | | Authors: | Chang, L, Li, Z, Zhang, H. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural Basis for the Inhibition of CRISPR-Cas12a by Anti-CRISPR Proteins.
Cell Host Microbe, 25, 2019
|
|
6NMD
 
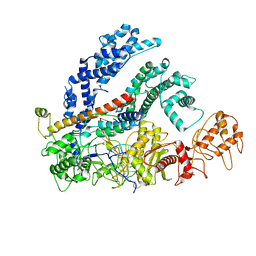 | | cryo-EM Structure of the LbCas12a-crRNA-AcrVA1 complex | | Descriptor: | AcrVA1, Cpf1, MAGNESIUM ION, ... | | Authors: | Chang, L, Li, Z, Zhang, H. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structural Basis for the Inhibition of CRISPR-Cas12a by Anti-CRISPR Proteins.
Cell Host Microbe, 25, 2019
|
|
6NM9
 
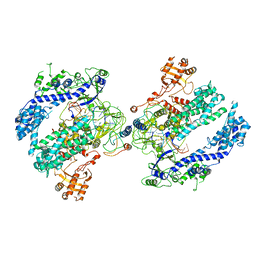 | | CryoEM structure of the LbCas12a-crRNA-AcrVA4 dimer | | Descriptor: | AcrVA4, Cpf1, MAGNESIUM ION, ... | | Authors: | Chang, L, Li, Z, Zhang, H. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural Basis for the Inhibition of CRISPR-Cas12a by Anti-CRISPR Proteins.
Cell Host Microbe, 25, 2019
|
|
6NME
 
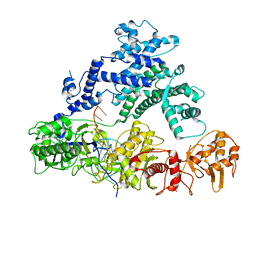 | | Structure of LbCas12a-crRNA | | Descriptor: | Cpf1, MAGNESIUM ION, crRNA | | Authors: | Chang, L, Li, Z, Zhang, H. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (5.67 Å) | | Cite: | Structural Basis for the Inhibition of CRISPR-Cas12a by Anti-CRISPR Proteins.
Cell Host Microbe, 25, 2019
|
|
6NMC
 
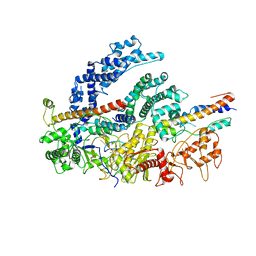 | | CryoEM structure of the LbCas12a-crRNA-2xAcrVA1 complex | | Descriptor: | AcrVA1, Cpf1, MAGNESIUM ION, ... | | Authors: | Chang, L, Li, Z, Zhang, H. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.24 Å) | | Cite: | Structural Basis for the Inhibition of CRISPR-Cas12a by Anti-CRISPR Proteins.
Cell Host Microbe, 25, 2019
|
|
6OMV
 
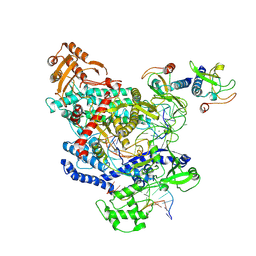 | | CryoEM structure of the LbCas12a-crRNA-AcrVA4-DNA complex | | Descriptor: | AcrVA4, Cpf1, DNA (5'-D(*CP*GP*TP*CP*CP*TP*TP*TP*AP*GP*GP*A)-3'), ... | | Authors: | Chang, L, Li, Z, Zhang, H. | | Deposit date: | 2019-04-19 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Basis for the Inhibition of CRISPR-Cas12a by Anti-CRISPR Proteins.
Cell Host Microbe, 25, 2019
|
|
6W62
 
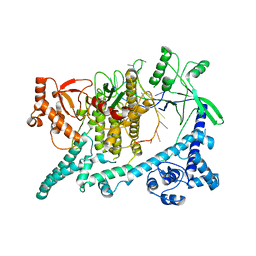 | | Cryo-EM structure of Cas12i-crRNA complex | | Descriptor: | Cas12i, crRNA | | Authors: | Chang, L, Li, Z, Zhang, H. | | Deposit date: | 2020-03-16 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mechanisms for target recognition and cleavage by the Cas12i RNA-guided endonuclease.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6W5C
 
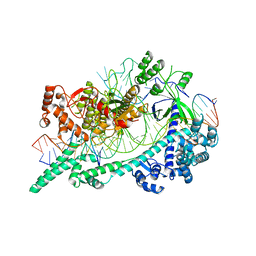 | | Cryo-EM structure of Cas12i(E894A)-crRNA-dsDNA complex | | Descriptor: | Cas12i, NTS, Substrate, ... | | Authors: | Chang, L, Li, Z, Zhang, H. | | Deposit date: | 2020-03-13 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanisms for target recognition and cleavage by the Cas12i RNA-guided endonuclease.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VYJ
 
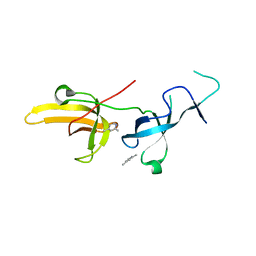 | | Human UHRF1 TTD domain in complex with a fragment | | Descriptor: | 2,4-dimethylpyridine, E3 ubiquitin-protein ligase UHRF1, beta-D-glucopyranose | | Authors: | Campbell, J.C, Chang, L, Young, D.W. | | Deposit date: | 2020-02-26 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Discovery of small molecules targeting the tandem tudor domain of the epigenetic factor UHRF1 using fragment-based ligand discovery.
Sci Rep, 11, 2021
|
|
6W64
 
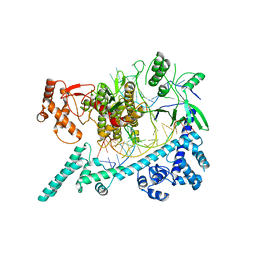 | |
6W92
 
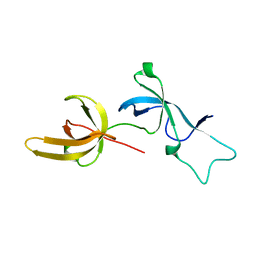 | | Human UHRF1 TTD domain | | Descriptor: | E3 ubiquitin-protein ligase UHRF1 | | Authors: | Campbell, J.C, Chang, L, Sankaran, B, Young, D.W. | | Deposit date: | 2020-03-21 | | Release date: | 2021-02-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Discovery of small molecules targeting the tandem tudor domain of the epigenetic factor UHRF1 using fragment-based ligand discovery.
Sci Rep, 11, 2021
|
|
6XMF
 
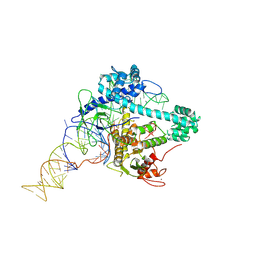 | |
6XMG
 
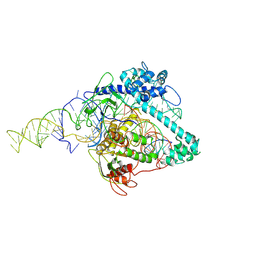 | | Cryo-EM structure of Cas12g ternary complex | | Descriptor: | CRISPR-Cas, RNA (130-MER), RNA (5'-R(P*UP*UP*AP*AP*UP*GP*CP*GP*GP*UP*AP*GP*UP*UP*UP*AP*UP*CP*AP*CP*AP*GP*UP*U)-3'), ... | | Authors: | Chang, L, Li, Z, Zhang, H. | | Deposit date: | 2020-06-30 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cryo-EM structure of the RNA-guided ribonuclease Cas12g.
Nat.Chem.Biol., 17, 2021
|
|
8FD2
 
 | | Cryo-EM structure of Cascade complex in type I-B CAST system | | Descriptor: | A Type I-B CRISPR-associated protein Cas5, RNA, Type I-B CRISPR-associated protein Cas11, ... | | Authors: | Chang, L, Wang, S. | | Deposit date: | 2022-12-01 | | Release date: | 2023-08-09 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Molecular mechanism for Tn7-like transposon recruitment by a type I-B CRISPR effector.
Cell, 186, 2023
|
|
