2WZB
 
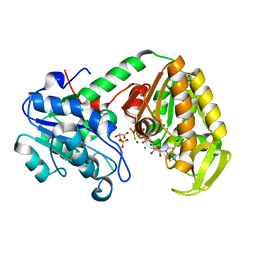 | | The catalytically active fully closed conformation of human phosphoglycerate kinase in complex with ADP, 3PG and magnesium trifluoride | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Bowler, M.W, Cliff, M.J, Marston, J.P.M, Baxter, N.J, Hownslow, A.M.H, Varga, A.V, Szabo, J, Vas, M, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-11-27 | | Release date: | 2010-04-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Transition State Analogue Structures of Human Phosphoglycerate Kinase Establish the Importance of Charge Balance in Catalysis.
J.Am.Chem.Soc., 132, 2010
|
|
5ADO
 
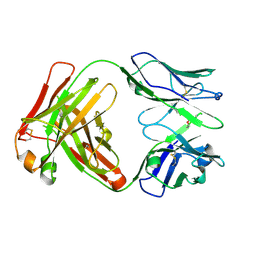 | | Crystal structure of the paraoxon-modified A.17 antibody FAB fragment - Light chain S35R mutant | | Descriptor: | DIETHYL PHOSPHONATE, FAB A.17 | | Authors: | Chatziefthimiou, S.D, Smirnov, I.V, Golovin, A.V, Stepanova, A.V, Peng, Y, Zolotareva, O.I, Belogurov, A.A, Ponomarenko, N.A, Blackburn, G.M, Gabibov, A.A, Lerner, R, Wilmanns, M. | | Deposit date: | 2015-08-21 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Robotic Qm/Mm-Driven Maturation of Antibody Combining Sites.
Sci.Adv., 2, 2016
|
|
5ADP
 
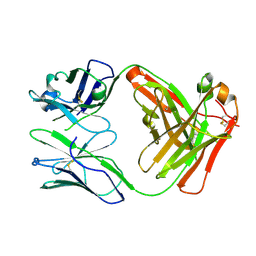 | | Crystal structure of the A.17 antibody FAB fragment - Light chain S35R mutant | | Descriptor: | FAB A.17 | | Authors: | Chatziefthimiou, S.D, Smirnov, I.V, Golovin, A.V, Stepanova, A.V, Peng, Y, Zolotareva, O.I, Belogurov, A.A, Ponomarenko, N.A, Blackburn, G.M, Gabibov, A.A, Lerner, R, Wilmanns, M. | | Deposit date: | 2015-08-21 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Robotic Qm/Mm-Driven Maturation of Antibody Combining Sites.
Sci.Adv., 2, 2016
|
|
3BL0
 
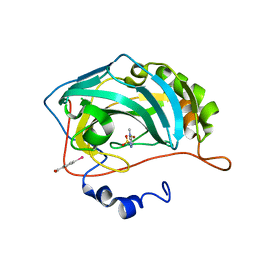 | | Carbonic anhydrase inhibitors. Interaction of 2-N,N-Dimethylamino-1,3,4-thiadiazole-5-methanesulfonamide with twelve mammalian isoforms: kinetic and X-Ray crystallographic studies | | Descriptor: | 1-[5-(dimethylamino)-1,3,4-thiadiazol-2-yl]methanesulfonamide, Carbonic anhydrase 2, MERCURIBENZOIC ACID, ... | | Authors: | Temperini, C, Supuran, C.T, Blackburn, G.M. | | Deposit date: | 2007-12-10 | | Release date: | 2008-01-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Carbonic anhydrase inhibitors. Interaction of 2-N,N-dimethylamino-1,3,4-thiadiazole-5-methanesulfonamide with 12 mammalian isoforms: kinetic and X-ray crystallographic studies.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
1RZY
 
 | | Crystal structure of rabbit Hint complexed with N-ethylsulfamoyladenosine | | Descriptor: | 5'-O-(N-ETHYL-SULFAMOYL)ADENOSINE, Histidine triad nucleotide-binding protein 1 | | Authors: | Krakowiak, A.K, Pace, H.C, Blackburn, G.M, Adams, M, Mekhalfia, A, Kaczmarek, R, Baraniak, J, Stec, W.J, Brenner, C. | | Deposit date: | 2003-12-29 | | Release date: | 2004-03-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biochemical, crystallographic, and mutagenic characterization of hint, the AMP-lysine hydrolase, with novel substrates and inhibitors
J.Biol.Chem., 279, 2004
|
|
2AIO
 
 | | Metallo beta lactamase L1 from Stenotrophomonas maltophilia complexed with hydrolyzed moxalactam | | Descriptor: | (2R)-2-((R)-CARBOXY{[CARBOXY(4-HYDROXYPHENYL)ACETYL]AMINO}METHOXYMETHYL)-5-METHYLENE-5,6-DIHYDRO-2H-1,3-OXAZINE-4-CARBO XYLIC ACID, Metallo-beta-lactamase L1, SULFATE ION, ... | | Authors: | Spencer, J, Read, J, Sessions, R.B, Howell, S, Blackburn, G.M, Gamblin, S.J. | | Deposit date: | 2005-07-30 | | Release date: | 2005-10-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Antibiotic Recognition by Binuclear Metallo-beta-Lactamases Revealed by X-ray Crystallography
J.Am.Chem.Soc., 127, 2005
|
|
2EU2
 
 | | Human Carbonic Anhydrase II in complex with novel inhibitors | | Descriptor: | (R)-1-AMINO-1-[5-(DIMETHYLAMINO)-1,3,4-THIADIAZOL-2-YL]METHANESULFONAMIDE, Carbonic anhydrase 2, ZINC ION | | Authors: | Fisher, S.Z, Govindasamy, L, Boyle, N, Agbandje-McKenna, M, Silverman, D.N, Blackburn, G.M, McKenna, R. | | Deposit date: | 2005-10-28 | | Release date: | 2006-07-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | X-ray crystallographic studies reveal that the incorporation of spacer groups in carbonic anhydrase inhibitors causes alternate binding modes.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
6RWZ
 
 | | Structure of Zika virus NS3 helicase in complex with ADP-BeF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MANGANESE (II) ION, ... | | Authors: | Ge, M, Molt Jr, R.W, Jenkins, H.T, Blackburn, G.M, Jin, Y, Antson, A.A. | | Deposit date: | 2019-06-07 | | Release date: | 2020-07-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Octahedral Trifluoromagnesate, an Anomalous Metal Fluoride Species, Stabilizes the Transition State in a Biological Motor.
Acs Catalysis, 11, 2021
|
|
6RVU
 
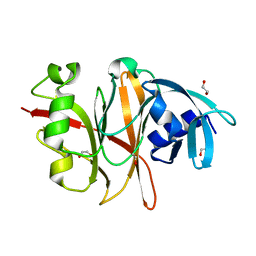 | | Crystal structure of the Burkholderia Lethal Factor 1 (BLF1) | | Descriptor: | 1,2-ETHANEDIOL, Lethal Factor 1 (BLF1) | | Authors: | Mobbs, G.W, Aziz, A.A, Blackburn, G.M, Sedelnikova, S.E, Minshull, T.C, Dickman, M.J, Baker, P.J, Nathan, S, Firdaus-Raih, M, Rice, D.W. | | Deposit date: | 2019-06-01 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Molecular basis of specificity and deamidation of eIF4A by Burkholderia Lethal Factor 1.
Commun Biol, 5, 2022
|
|
6S0J
 
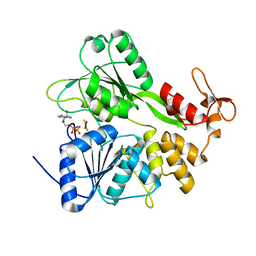 | | Structure of Zika virus NS3 helicase in complex with ADP-MgF3(H2O)- | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Genome polyprotein, MAGNESIUM ION, ... | | Authors: | Ge, M, Molt Jr, R.W, Jenkins, H.T, Blackburn, G.M, Jin, Y, Antson, A.A. | | Deposit date: | 2019-06-16 | | Release date: | 2020-07-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Octahedral Trifluoromagnesate, an Anomalous Metal Fluoride Species, Stabilizes the Transition State in a Biological Motor.
Acs Catalysis, 11, 2021
|
|
7PQ0
 
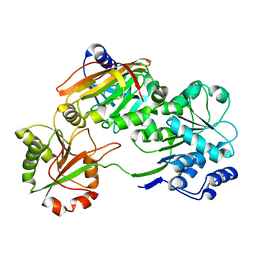 | | Crystal structure of the Burkholderia Lethal Factor 1 (BLF1) C94S inactive mutant in complex with human eIF4A - Crystal form B | | Descriptor: | Burkholderia Lethal Factor 1 (BLF1), Eukaryotic initiation factor 4A-I | | Authors: | Mobbs, G.W, Aziz, A.A, Dix, S.R, Blackburn, G.M, Sedelnikova, S.E, Minshull, T.C, Dickman, M.J, Baker, P.J, Nathan, S, Firdaus-Raih, M, Rice, D.W. | | Deposit date: | 2021-09-15 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular basis of specificity and deamidation of eIF4A by Burkholderia Lethal Factor 1.
Commun Biol, 5, 2022
|
|
7PPZ
 
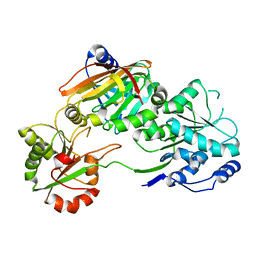 | | Crystal structure of the Burkholderia Lethal Factor 1 (BLF1) C94S inactive mutant in complex with human eIF4A - Crystal form A | | Descriptor: | Burkholderia Lethal Factor 1 (BLF1), Eukaryotic initiation factor 4A-I | | Authors: | Mobbs, G.W, Aziz, A.A, Dix, S.R, Blackburn, G.M, Sedelnikova, S.E, Minshull, T.C, Dickman, M.J, Baker, P.J, Nathan, S, Firdaus-Raih, M, Rice, D.W. | | Deposit date: | 2021-09-15 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Molecular basis of specificity and deamidation of eIF4A by Burkholderia Lethal Factor 1.
Commun Biol, 5, 2022
|
|
1X8E
 
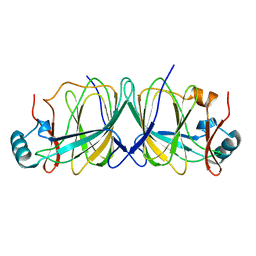 | | Crystal structure of Pyrococcus furiosus phosphoglucose isomerase free enzyme | | Descriptor: | Glucose-6-phosphate isomerase | | Authors: | Berrisford, J.M, Akerboom, J, Brouns, S, Sedelnikova, S.E, Turnbull, A.P, van der Oost, J, Salmon, L, Hardre, R, Murray, I.A, Blackburn, G.M, Rice, D.W, Baker, P.J. | | Deposit date: | 2004-08-18 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structures of inhibitor complexes of Pyrococcus furiosus phosphoglucose isomerase provide insights into substrate binding and catalysis.
J.Mol.Biol., 343, 2004
|
|
1X82
 
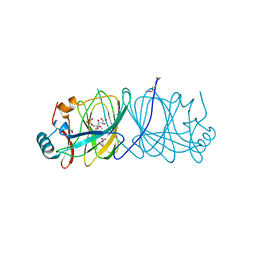 | | CRYSTAL STRUCTURE OF PHOSPHOGLUCOSE ISOMERASE FROM PYROCOCCUS FURIOSUS WITH BOUND 5-phospho-D-arabinonate | | Descriptor: | 5-PHOSPHOARABINONIC ACID, Glucose-6-phosphate isomerase | | Authors: | Berrisford, J.M, Akerboom, J, Brouns, S, Sedelnikova, S.E, Turnbull, A.P, van der Oost, J, Salmon, L, Hardre, R, Murray, I.A, Blackburn, G.M, Rice, D.W, Baker, P.J. | | Deposit date: | 2004-08-17 | | Release date: | 2004-10-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structures of inhibitor complexes of Pyrococcus furiosus phosphoglucose isomerase provide insights into substrate binding and catalysis.
J.Mol.Biol., 343, 2004
|
|
1X7N
 
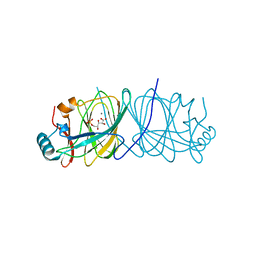 | | The crystal structure of Pyrococcus furiosus phosphoglucose isomerase with bound 5-phospho-D-arabinonate and Manganese | | Descriptor: | 5-PHOSPHOARABINONIC ACID, Glucose-6-phosphate isomerase, MANGANESE (II) ION | | Authors: | Berrisford, J.M, Akerboom, J, Brouns, S, Sedelnikova, S.E, Turnbull, A.P, van der Oost, J, Salmon, L, Hardre, R, Murray, I.A, Blackburn, G.M, Rice, D.W, Baker, P.J. | | Deposit date: | 2004-08-16 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The structures of inhibitor complexes of Pyrococcus furiosus phosphoglucose isomerase provide insights into substrate binding and catalysis.
J.Mol.Biol., 343, 2004
|
|
2WF9
 
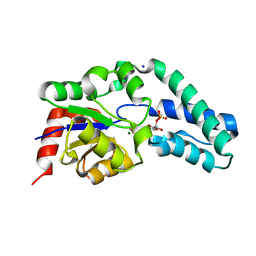 | | Structure of Beta-Phosphoglucomutase inhibited with Glucose-6- phosphate, and Beryllium trifluoride, crystal form 2 | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, 6-O-phosphono-beta-D-glucopyranose, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Bowler, M.W, Baxter, N.J, Webster, C.E, Pollard, S, Alizadeh, T, Hounslow, A.M, Cliff, M.J, Bermel, W, Williams, N.H, Hollfelder, F, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-04-03 | | Release date: | 2010-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Near Attack Conformers Dominate Beta-Phosphoglucomutase Complexes Where Geometry and Charge Distribution Reflect Those of Substrate.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2WF8
 
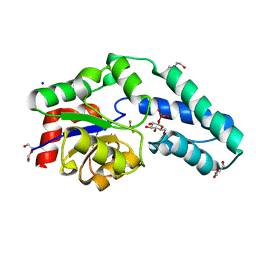 | | Structure of Beta-Phosphoglucomutase inhibited with Glucose-6- phosphate, Glucose-1-phosphate and Beryllium trifluoride | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, 6-O-phosphono-beta-D-glucopyranose, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Bowler, M.W, Baxter, N.J, Webster, C.E, Pollard, S, Alizadeh, T, Hounslow, A.M, Cliff, M.J, Bermel, W, Williams, N.H, Hollfelder, F, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-04-03 | | Release date: | 2010-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Near attack conformers dominate beta-phosphoglucomutase complexes where geometry and charge distribution reflect those of substrate.
Proc. Natl. Acad. Sci. U.S.A., 109, 2012
|
|
2WFA
 
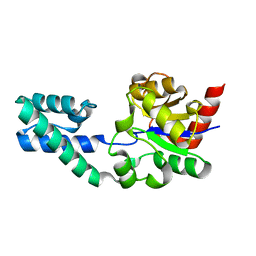 | | Structure of Beta-Phosphoglucomutase inhibited with Beryllium trifluoride, in an open conformation. | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, BETA-PHOSPHOGLUCOMUTASE, MAGNESIUM ION | | Authors: | Bowler, M.W, Baxter, N.J, Webster, C.E, Pollard, S, Alizadeh, T, Hounslow, A.M, Cliff, M.J, Bermel, W, Williams, N.H, Hollfelder, F, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-04-03 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Near Attack Conformers Dominate Beta-Phosphoglucomutase Complexes Where Geometry and Charge Distribution Reflect Those of Substrate.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2WF7
 
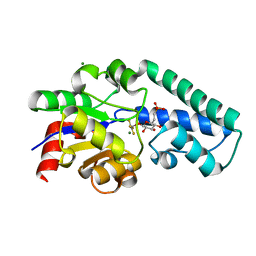 | | Structure of Beta-Phosphoglucomutase inhibited with Glucose-6- phosphonate and Aluminium tetrafluoride | | Descriptor: | 6,7-dideoxy-7-phosphono-beta-D-gluco-heptopyranose, BETA-PHOSPHOGLUCOMUTASE, MAGNESIUM ION, ... | | Authors: | Bowler, M.W, Baxter, N.J, Webster, C.E, Pollard, S, Alizadeh, T, Hounslow, A.M, Cliff, M.J, Bermel, W, Williams, N.H, Hollfelder, F, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-04-03 | | Release date: | 2010-05-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Alpha-Fluorophosphonates Reveal How a Phosphomutase Conserves Transition State Conformation Over Hexose Recognition in its Two-Step Reaction.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2WF6
 
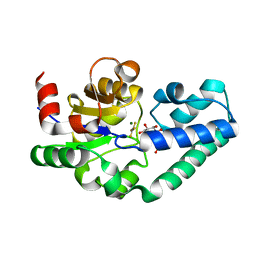 | | Structure of Beta-Phosphoglucomutase inhibited with Glucose-6-phosphate and Aluminium tetrafluoride | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, BETA-PHOSPHOGLUCOMUTASE, MAGNESIUM ION, ... | | Authors: | Bowler, M.W, Baxter, N.J, Webster, C.E, Pollard, S, Alizadeh, T, Hounslow, A.M, Cliff, M.J, Bermel, W, Williams, N.H, Hollfelder, F, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-04-03 | | Release date: | 2010-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Atomic details of near-transition state conformers for enzyme phosphoryl transfer revealed by MgF-3 rather than by phosphoranes.
Proc. Natl. Acad. Sci. U.S.A., 107, 2010
|
|
2WF5
 
 | | Structure of Beta-Phosphoglucomutase inhibited with Glucose-6-phosphate and trifluoromagnesate | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, BETA-PHOSPHOGLUCOMUTASE, MAGNESIUM ION, ... | | Authors: | Bowler, M.W, Baxter, N.J, Webster, C.E, Hounslow, A.M, Cliff, M.J, Williams, N.H, Hollfelder, F, Gamblin, S, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-04-03 | | Release date: | 2009-09-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Atomic details of near-transition state conformers for enzyme phosphoryl transfer revealed by MgF-3 rather than by phosphoranes.
Proc. Natl. Acad. Sci. U.S.A., 107, 2010
|
|
2WHE
 
 | | Structure of native Beta-Phosphoglucomutase in an open conformation without bound ligands. | | Descriptor: | BETA-PHOSPHOGLUCOMUTASE, MAGNESIUM ION | | Authors: | Bowler, M.W, Baxter, N.J, Webster, C.E, Pollard, S, Alizadeh, T, Hounslow, A.M, Cliff, M.J, Bermel, W, Williams, N.H, Hollfelder, F, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-05-04 | | Release date: | 2009-09-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Atomic Details of Near-Transition State Conformers for Enzyme Phosphoryl Transfer Revealed by Mgf-3 Rather Than by Phosphoranes.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2WZD
 
 | | The catalytically active fully closed conformation of human phosphoglycerate kinase K219A mutant in complex with ADP, 3PG and aluminium trifluoride | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, ... | | Authors: | Bowler, M.W, Cliff, M.J, Marston, J.P.M, Baxter, N.J, Hownslow, A.M.H, Varga, A.V, Szabo, J, Vas, M, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-11-27 | | Release date: | 2010-04-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Transition State Analogue Structures of Human Phosphoglycerate Kinase Establish the Importance of Charge Balance in Catalysis.
J.Am.Chem.Soc., 132, 2010
|
|
2X15
 
 | | The catalytically active fully closed conformation of human phosphoglycerate kinase in complex with ADP and 1,3- bisphosphoglycerate | | Descriptor: | 1,3-BISPHOSPHOGLYCERIC ACID, 3-PHOSPHOGLYCERIC ACID, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Bowler, M.W, Cliff, M.J, Marston, J.P.M, Baxter, N.J, Hounslow, A.M.H, Varga, A.V, Szabo, J, Vas, M, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-12-21 | | Release date: | 2011-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of Human Phosphoglycerate Kinase in its Fully Active Conformation in Complex with Ground State Analoges
To be Published
|
|
2WZC
 
 | | The catalytically active fully closed conformation of human phosphoglycerate kinase in complex with ADP, 3PG and aluminium tetrafluoride | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Bowler, M.W, Cliff, M.J, Marston, J.P.M, Baxter, N.J, Hownslow, A.M.H, Varga, A.V, Szabo, J, Vas, M, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-11-27 | | Release date: | 2010-04-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Transition State Analogue Structures of Human Phosphoglycerate Kinase Establish the Importance of Charge Balance in Catalysis.
J.Am.Chem.Soc., 132, 2010
|
|
