6GCG
 
 | | Copper nitrite reductase from Achromobacter cycloclastes: large polymorph dataset 15 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase | | Authors: | Ebrahim, A, Appleby, M.V, Axford, D, Beale, J, Moreno-Chicano, T, Sherrell, D.A, Strange, R.W, Owen, R.L, Hough, M.A. | | Deposit date: | 2018-04-17 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.80015242 Å) | | Cite: | Resolving polymorphs and radiation-driven effects in microcrystals using fixed-target serial synchrotron crystallography.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
4GV1
 
 | | PKB alpha in complex with AZD5363 | | Descriptor: | 4-amino-N-[(1S)-1-(4-chlorophenyl)-3-hydroxypropyl]-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidine-4-carboxamide, GLYCEROL, RAC-alpha serine/threonine-protein kinase | | Authors: | Addie, M, Ballard, P, Bird, G, Buttar, D, Currie, G, Davies, B, Debreczeni, J, Dry, H, Dudley, P, Greenwood, R, Hatter, G, Jestel, A, Johnson, P.D, Kettle, J.G, Lane, C, Lamont, G, Leach, A, Luke, R.W.A, Ogilvie, D, Page, K, Pass, M, Steinbacher, S, Steuber, H, Pearson, S, Ruston, L. | | Deposit date: | 2012-08-30 | | Release date: | 2013-02-27 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Discovery of 4-Amino-N-[(1S)-1-(4-chlorophenyl)-3-hydroxypropyl]-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidine-4-carboxamide (AZD5363), an Orally Bioavailable, Potent Inhibitor of Akt Kinases.
J.Med.Chem., 56, 2013
|
|
6ZFV
 
 | |
5EIV
 
 | | Crystal structure of complex of osteoclast-associated immunoglobulin-like receptor (OSCAR) and a synthetic collagen consensus peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CHLORIDE ION, ... | | Authors: | Zhou, L, Blaszczyk, M, Chirgadze, D, Bihan, D, Farndale, R.W. | | Deposit date: | 2015-10-30 | | Release date: | 2015-11-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.414 Å) | | Cite: | Structural basis for collagen recognition by the immune receptor OSCAR.
Blood, 127, 2016
|
|
8T1O
 
 | | AP2 bound to MSP2N2 nanodisc with Tgn38 cargo peptide; composite map | | Descriptor: | AP-2 complex subunit alpha-2, AP-2 complex subunit beta, AP-2 complex subunit mu, ... | | Authors: | Baker, R.W, Cannon, K.S, Reta, S. | | Deposit date: | 2023-06-02 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Lipid nanodiscs as a template for high-resolution cryo-EM structures of peripheral membrane proteins.
J.Struct.Biol., 215, 2023
|
|
4WFW
 
 | |
7YTJ
 
 | | Cryo-EM structure of VTC complex | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, INOSITOL HEXAKISPHOSPHATE, PHOSPHATE ION, ... | | Authors: | Guan, Z.Y, Chen, J, Liu, R.W, Chen, Y.K, Xing, Q, Du, Z.M, Liu, Z. | | Deposit date: | 2022-08-15 | | Release date: | 2023-02-22 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The cytoplasmic synthesis and coupled membrane translocation of eukaryotic polyphosphate by signal-activated VTC complex.
Nat Commun, 14, 2023
|
|
5O9B
 
 | |
6TKA
 
 | | Crystal structure of human O-GlcNAc transferase bound to substrate 7 and a peptide from HCF-1 pro-repeat 2 (11-26) | | Descriptor: | 3-[2,2-bis(fluoranyl)-10,12-dimethyl-1,3-diaza-2$l^{4}-boratricyclo[7.3.0.0^{3,7}]dodeca-4,6,9,11-tetraen-4-yl]-~{N}-ethyl-propanamide, HCF-1 pro-repeat 2 (11-26), UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit, ... | | Authors: | Meek, R.W, Davies, G.J. | | Deposit date: | 2019-11-28 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | A Direct Fluorescent Activity Assay for Glycosyltransferases Enables Convenient High-Throughput Screening: Application to O-GlcNAc Transferase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
8FB5
 
 | | Crystal structure of Ky15.11-S100IK Fab in complex with circumsporozoite protein KQPA peptide | | Descriptor: | Circumsporozoite protein KQPA peptide, Ky15.11-SK Antibody, heavy chain, ... | | Authors: | Kang, R.W, Thai, E, Julien, J.P. | | Deposit date: | 2022-11-29 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular determinants of cross-reactivity and potency by VH3-33 antibodies against the Plasmodium falciparum circumsporozoite protein.
Cell Rep, 42, 2023
|
|
8FB8
 
 | | Crystal structure of Ky15.10-Y100EK Fab in complex with circumsporozoite protein KQPA peptide | | Descriptor: | Circumsporozoite protein KQPA peptide, GLYCEROL, Ky15.10-YK Antibody, ... | | Authors: | Kang, R.W, Thai, E, Julien, J.P. | | Deposit date: | 2022-11-29 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Molecular determinants of cross-reactivity and potency by VH3-33 antibodies against the Plasmodium falciparum circumsporozoite protein.
Cell Rep, 42, 2023
|
|
1IRA
 
 | | COMPLEX OF THE INTERLEUKIN-1 RECEPTOR WITH THE INTERLEUKIN-1 RECEPTOR ANTAGONIST (IL1RA) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, INTERLEUKIN-1 RECEPTOR, INTERLEUKIN-1 RECEPTOR ANTAGONIST | | Authors: | Schreuder, H.A, Tardif, C, Tramp-Kalmeyer, S, Soffientini, A, Sarubbi, E, Akeson, A, Bowlin, T, Yanofsky, S, Barrett, R.W. | | Deposit date: | 1998-04-09 | | Release date: | 1998-06-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A new cytokine-receptor binding mode revealed by the crystal structure of the IL-1 receptor with an antagonist.
Nature, 386, 1997
|
|
5WEC
 
 | |
1WRP
 
 | | FLEXIBILITY OF THE DNA-BINDING DOMAINS OF TRP REPRESSOR | | Descriptor: | TRP REPRESSOR, TRYPTOPHAN | | Authors: | Schewitz, R.W, Otwinowski, Z, Lawson, C.L, Joachimiak, A, Sigler, P.B. | | Deposit date: | 1987-12-01 | | Release date: | 1988-04-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Flexibility of the DNA-binding domains of trp repressor.
Proteins, 3, 1988
|
|
6TNA
 
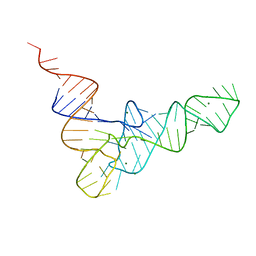 | | CRYSTAL STRUCTURE OF YEAST PHENYLALANINE T-RNA. I.CRYSTALLOGRAPHIC REFINEMENT | | Descriptor: | MAGNESIUM ION, TRNAPHE | | Authors: | Sussman, J.L, Holbrook, S.R, Warrant, R.W, Church, G.M, Kim, S.-H. | | Deposit date: | 1978-11-16 | | Release date: | 1979-01-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of yeast phenylalanine transfer RNA. I. Crystallographic refinement.
J.Mol.Biol., 123, 1978
|
|
1IF0
 
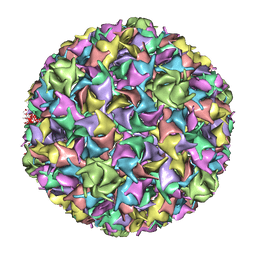 | | PSEUDO-ATOMIC MODEL OF BACTERIOPHAGE HK97 PROCAPSID (PROHEAD II) | | Descriptor: | PROTEIN (MAJOR CAPSID PROTEIN GP5) | | Authors: | Conway, J.F, Wikoff, W.R, Cheng, N, Duda, R.L, Hendrix, R.W, Johnson, J.E, Steven, A.C. | | Deposit date: | 2001-04-11 | | Release date: | 2001-05-02 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Virus maturation involving large subunit rotations and local refolding.
Science, 292, 2001
|
|
5W8O
 
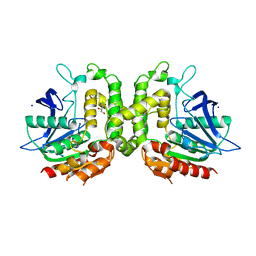 | | Homoserine transacetylase MetX from Mycobacterium hassiacum | | Descriptor: | CALCIUM ION, GLYCEROL, Homoserine O-acetyltransferase, ... | | Authors: | Reed, R.W, Rodriguez, E.S, Li, J, Korotkov, K.V. | | Deposit date: | 2017-06-22 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural analysis of mycobacterial homoserine transacetylases central to methionine biosynthesis reveals druggable active site.
Sci Rep, 9, 2019
|
|
1XEU
 
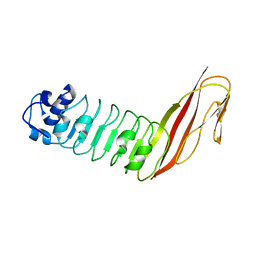 | | Crystal Structure of Internalin C from Listeria monocytogenes | | Descriptor: | internalin C | | Authors: | Ooi, A, Hussain, S, Seyedarabi, A, Pickersgill, R.W. | | Deposit date: | 2004-09-13 | | Release date: | 2005-08-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of internalin C from Listeria monocytogenes.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
6MQ4
 
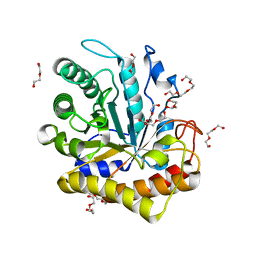 | | GH5-4 broad specificity endoglucanase from Hungateiclostridium cellulolyticum | | Descriptor: | DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL, ... | | Authors: | Bingman, C.A, Smith, R.W, Glasgow, E.M, Fox, B.G. | | Deposit date: | 2018-10-09 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis.
J.Biol.Chem., 295, 2020
|
|
6N64
 
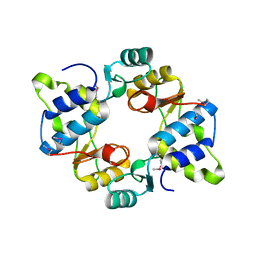 | | Crystal structure of mouse SMCHD1 hinge domain | | Descriptor: | Structural maintenance of chromosomes flexible hinge domain-containing protein 1, Uncharacterized peptide from Structural maintenance of chromosomes flexible hinge domain-containing protein 1 | | Authors: | Birkinshaw, R.W, Chen, K, Czabotar, P.E, Blewitt, M.E, Murphy, J.M. | | Deposit date: | 2018-11-25 | | Release date: | 2020-06-17 | | Last modified: | 2021-01-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the hinge domain of Smchd1 reveals its dimerization mode and nucleic acid-binding residues.
Sci.Signal., 13, 2020
|
|
1E5N
 
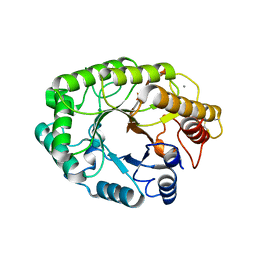 | | E246C mutant of P fluorescens subsp. cellulosa xylanase A in complex with xylopentaose | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE A, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Lo Leggio, L, Jenkins, J.A, Harris, G.W, Pickersgill, R.W. | | Deposit date: | 2000-07-27 | | Release date: | 2000-12-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray crystallographic study of xylopentaose binding to Pseudomonas fluorescens xylanase A.
Proteins, 41, 2000
|
|
4X6E
 
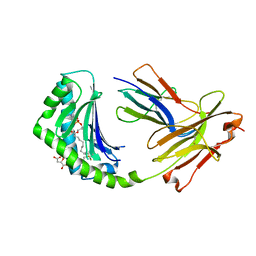 | | CD1a binary complex with lysophosphatidylcholine | | Descriptor: | (4R,7R,18Z)-4,7-dihydroxy-N,N,N-trimethyl-10-oxo-3,5,9-trioxa-4-phosphaheptacos-18-en-1-aminium 4-oxide, Beta-2-microglobulin, D-MALATE, ... | | Authors: | Birkinshaw, R.W, Rossjohn, J. | | Deposit date: | 2014-12-08 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | alpha beta T cell antigen receptor recognition of CD1a presenting self lipid ligands.
Nat.Immunol., 16, 2015
|
|
3CZW
 
 | | X-ray structures of the (GUGGUCUGAUGAGGCC) RNA duplex | | Descriptor: | RNA (5'-R(*G*UP*GP*GP*UP*CP*UP*GP*AP*UP*GP*AP*GP*GP*CP*C)-3'), SULFATE ION | | Authors: | Rypniewski, W, Adamiak, D.A, Milecki, J, Adamiak, R.W. | | Deposit date: | 2008-04-30 | | Release date: | 2008-09-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Noncanonical G(syn)-G(anti) base pairs stabilized by sulphate anions in two X-ray structures of the (GUGGUCUGAUGAGGCC) RNA duplex.
Rna, 14, 2008
|
|
4X6F
 
 | | CD1a binary complex with sphingomyelin | | Descriptor: | (4S,7S,23Z)-4-hydroxy-7-[(1S,2Z)-1-hydroxyhexadec-2-en-1-yl]-N,N,N-trimethyl-9-oxo-3,5-dioxa-8-aza-4-phosphadotriacont- 23-en-1-aminium 4-oxide, Beta-2-microglobulin, T-cell surface glycoprotein CD1a | | Authors: | Birkinshaw, R.W, Rossjohn, J. | | Deposit date: | 2014-12-08 | | Release date: | 2015-02-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | alpha beta T cell antigen receptor recognition of CD1a presenting self lipid ligands.
Nat.Immunol., 16, 2015
|
|
4X7G
 
 | | CobK precorrin-6A reductase | | Descriptor: | 3-[(1R,2S,3S,7S,11S,16S,17R,18R,19R)-2,7,12,18-tetrakis(2-hydroxy-2-oxoethyl)-3,13-bis(3-hydroxy-3-oxopropyl)-1,2,5,7,11,17-hexamethyl-17-(3-oxidanyl-3-oxidanylidene-prop-1-enyl)-3,6,8,10,15,16,18,19,21,24-decahydrocorrin-8-yl]propanoic acid, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Precorrin-6A reductase | | Authors: | Gu, S, Deery, E, Warren, M.J, Pickersgill, R.W. | | Deposit date: | 2014-12-09 | | Release date: | 2016-01-20 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Crystal structure of CobK reveals strand-swapping between Rossmann-fold domains and molecular basis of the reduced precorrin product trap.
Sci Rep, 5, 2015
|
|
