1IYV
 
 | |
1IYU
 
 | |
1GHK
 
 | | SOLUTION STRUCTURE OF THE LIPOYL DOMAIN OF THE 2-OXOGLUTARATE DEHYDROGENASE COMPLEX FROM AZOTOBACTER VINELAND II, NMR, 25 STRUCTURES | | Descriptor: | E2, THE DIHYDROLIPOAMIDE SUCCINYLTRANSFERASE COMPONENT OF 2-OXOGLUTARATE DEHYDROGENASE COMPLEX | | Authors: | Berg, A, Vervoort, J, De Kok, A. | | Deposit date: | 1996-01-16 | | Release date: | 1997-01-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the lipoyl domain of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii.
J.Mol.Biol., 261, 1996
|
|
1GHJ
 
 | | SOLUTION STRUCTURE OF THE LIPOYL DOMAIN OF THE 2-OXOGLUTARATE DEHYDROGENASE COMPLEX FROM AZOTOBACTER VINELAND II, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | E2, THE DIHYDROLIPOAMIDE SUCCINYLTRANSFERASE COMPONENT OF 2-OXOGLUTARATE DEHYDROGENASE COMPLEX | | Authors: | Berg, A, Vervoort, J, De Kok, A. | | Deposit date: | 1996-01-16 | | Release date: | 1997-01-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the lipoyl domain of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii.
J.Mol.Biol., 261, 1996
|
|
7S89
 
 | | Open apo-state cryo-EM structure of human TRPV6 in cNW11 nanodiscs | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, ... | | Authors: | Neuberger, A, Nadezhdin, K.D, Sobolevsky, A.I. | | Deposit date: | 2021-09-17 | | Release date: | 2021-11-17 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Structural mechanisms of TRPV6 inhibition by ruthenium red and econazole.
Nat Commun, 12, 2021
|
|
7S8C
 
 | | Cryo-EM structure of human TRPV6 in complex with inhibitor econazole | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1-[(2R)-2-[(4-chlorobenzyl)oxy]-2-(2,4-dichlorophenyl)ethyl]-1H-imidazole, CALCIUM ION, ... | | Authors: | Neuberger, A, Nadezhdin, K.D, Sobolevsky, A.I. | | Deposit date: | 2021-09-17 | | Release date: | 2021-11-17 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural mechanisms of TRPV6 inhibition by ruthenium red and econazole.
Nat Commun, 12, 2021
|
|
7S88
 
 | | Open apo-state cryo-EM structure of human TRPV6 in glyco-diosgenin detergent | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, ... | | Authors: | Neuberger, A, Nadezhdin, K.D, Sobolevsky, A.I. | | Deposit date: | 2021-09-17 | | Release date: | 2021-11-17 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structural mechanisms of TRPV6 inhibition by ruthenium red and econazole.
Nat Commun, 12, 2021
|
|
7ZQ9
 
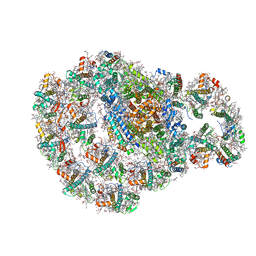 | | Dimeric PSI of Chlamydomonas reinhardtii at 2.74 A resolution (symmetry expanded) | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Naschberger, A, Amunts, A. | | Deposit date: | 2022-04-29 | | Release date: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Algal photosystem I dimer and high-resolution model of PSI-plastocyanin complex.
Nat.Plants, 8, 2022
|
|
7ZQC
 
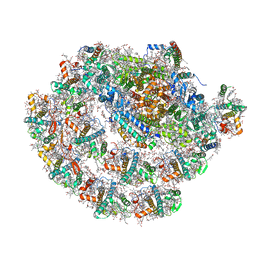 | | Monomeric PSI of Chlamydomonas reinhardtii at 2.31 A resolution | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Naschberger, A, Amunts, A. | | Deposit date: | 2022-04-29 | | Release date: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.31 Å) | | Cite: | Algal photosystem I dimer and high-resolution model of PSI-plastocyanin complex.
Nat.Plants, 8, 2022
|
|
7ZQE
 
 | |
7ZQD
 
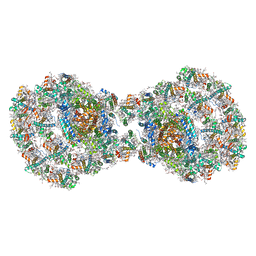 | | Dimeric PSI of Chlamydomonas reinhardtii at 2.97 A resolution | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Naschberger, A, Amunts, A. | | Deposit date: | 2022-04-29 | | Release date: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Algal photosystem I dimer and high-resolution model of PSI-plastocyanin complex.
Nat.Plants, 8, 2022
|
|
2XK0
 
 | | Solution structure of the Tudor domain from Drosophila Polycomblike (Pcl) | | Descriptor: | POLYCOMB PROTEIN PCL | | Authors: | Friberg, A, Oddone, A, Klymenko, T, Mueller, J, Sattler, M. | | Deposit date: | 2010-07-07 | | Release date: | 2010-08-11 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure of an Atypical Tudor Domain in the Drosophila Polycomblike Protein
Protein Sci., 19, 2010
|
|
7OS2
 
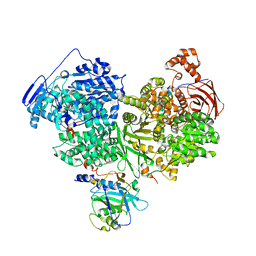 | | Cryo-EM structure of Brr2 in complex with Jab1/MPN and C9ORF78 | | Descriptor: | Pre-mRNA-processing-splicing factor 8, Telomere length and silencing protein 1 homolog, U5 small nuclear ribonucleoprotein 200 kDa helicase | | Authors: | Bergfort, A, Hilal, T, Weber, G, Wahl, M.C. | | Deposit date: | 2021-06-07 | | Release date: | 2022-02-23 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | The intrinsically disordered TSSC4 protein acts as a helicase inhibitor, placeholder and multi-interaction coordinator during snRNP assembly and recycling.
Nucleic Acids Res., 50, 2022
|
|
7OS1
 
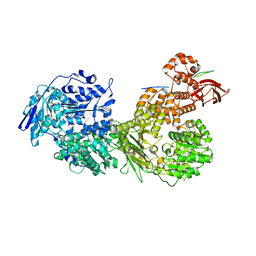 | | Cryo-EM structure of Brr2 in complex with Fbp21 | | Descriptor: | U5 small nuclear ribonucleoprotein 200 kDa helicase, WW domain-binding protein 4 | | Authors: | Bergfort, A, Hilal, T, Weber, G, Wahl, M.C. | | Deposit date: | 2021-06-07 | | Release date: | 2022-02-23 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The intrinsically disordered TSSC4 protein acts as a helicase inhibitor, placeholder and multi-interaction coordinator during snRNP assembly and recycling.
Nucleic Acids Res., 50, 2022
|
|
7PGU
 
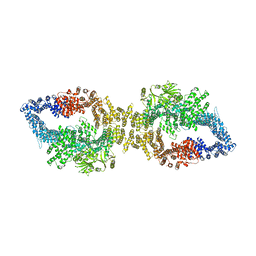 | | Autoinhibited structure of human neurofibromin isoform 2 stabilized by Zinc. | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, Neurofibromin, ZINC ION | | Authors: | Naschberger, A, Baradaran, R, Carroni, M, Rupp, B. | | Deposit date: | 2021-08-15 | | Release date: | 2021-11-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The structure of neurofibromin isoform 2 reveals different functional states.
Nature, 599, 2021
|
|
7PGT
 
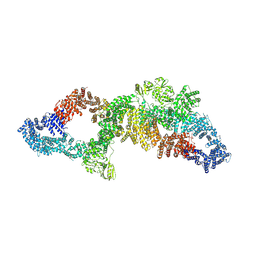 | | The structure of human neurofibromin isoform 2 in opened conformation. | | Descriptor: | Neurofibromin, ZINC ION | | Authors: | Naschberger, A, Baradaran, R, Carroni, M, Rupp, B. | | Deposit date: | 2021-08-15 | | Release date: | 2021-11-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | The structure of neurofibromin isoform 2 reveals different functional states.
Nature, 599, 2021
|
|
7PGS
 
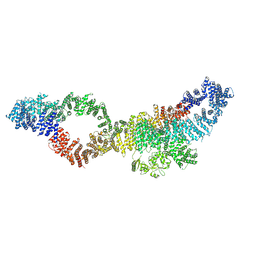 | | Consensus structure of human Neurofibromin isoform 2 | | Descriptor: | Neurofibromin, ZINC ION | | Authors: | Naschberger, A, Baradaran, R, Carroni, M, Rupp, B. | | Deposit date: | 2021-08-15 | | Release date: | 2021-11-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The structure of neurofibromin isoform 2 reveals different functional states.
Nature, 599, 2021
|
|
7PGR
 
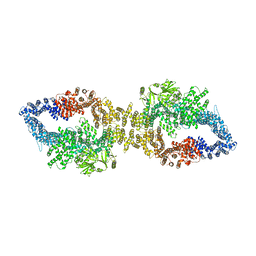 | | The structure of human neurofibromin isoform 2 in closed conformation | | Descriptor: | Neurofibromin, ZINC ION | | Authors: | Naschberger, A, Baradaran, R, Carroni, M, Rupp, B. | | Deposit date: | 2021-08-15 | | Release date: | 2021-11-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The structure of neurofibromin isoform 2 reveals different functional states.
Nature, 599, 2021
|
|
8FOB
 
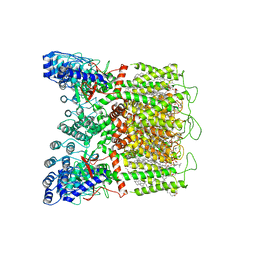 | | Cryo-EM structure of human TRPV6 in the open state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, ... | | Authors: | Neuberger, A, Yelshanskaya, M.V, Nadezhdin, K.D, Sobolevsky, A.I. | | Deposit date: | 2022-12-30 | | Release date: | 2023-05-24 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Structural mechanism of human oncochannel TRPV6 inhibition by the natural phytoestrogen genistein.
Nat Commun, 14, 2023
|
|
8FOA
 
 | | Cryo-EM structure of human TRPV6 in complex with the natural phytoestrogen genistein | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CALCIUM ION, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Neuberger, A, Yelshanskaya, M.V, Nadezhdin, K.D, Sobolevsky, A.I. | | Deposit date: | 2022-12-30 | | Release date: | 2023-05-24 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural mechanism of human oncochannel TRPV6 inhibition by the natural phytoestrogen genistein.
Nat Commun, 14, 2023
|
|
7UGG
 
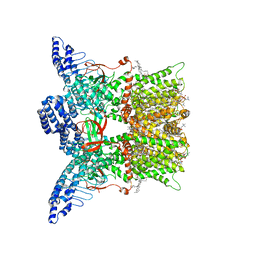 | | Cryo-EM structure of TRPV3 in complex with the anesthetic dyclonine | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Dyclonine, SODIUM ION, ... | | Authors: | Neuberger, A, Nadezhdin, K.D, Sobolevsky, A.I. | | Deposit date: | 2022-03-24 | | Release date: | 2022-06-08 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural mechanism of TRPV3 channel inhibition by the anesthetic dyclonine.
Nat Commun, 13, 2022
|
|
8GF8
 
 | | Cryo-EM structure of human TRPV1 in cNW11 nanodisc and soybean lipids | | Descriptor: | (2R)-3-{[(R)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctadecanoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, SODIUM ION, ... | | Authors: | Neuberger, A, Nadezhdin, K.D, Sobolevsky, A.I. | | Deposit date: | 2023-03-07 | | Release date: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Human TRPV1 structure and inhibition by the analgesic SB-366791.
Nat Commun, 14, 2023
|
|
8GF9
 
 | | Cryo-EM structure of human TRPV1 in cNW11 nanodisc and POPC:POPE:POPG lipids | | Descriptor: | (2R)-3-{[(R)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctadecanoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, SODIUM ION, ... | | Authors: | Neuberger, A, Nadezhdin, K.D, Sobolevsky, A.I. | | Deposit date: | 2023-03-07 | | Release date: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Human TRPV1 structure and inhibition by the analgesic SB-366791.
Nat Commun, 14, 2023
|
|
8GFA
 
 | | Cryo-EM structure of human TRPV1 in complex with the analgesic drug SB-366791 | | Descriptor: | (2E)-3-(4-chlorophenyl)-N-(3-methoxyphenyl)prop-2-enamide, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, ... | | Authors: | Neuberger, A, Nadezhdin, K.D, Sobolevsky, A.I. | | Deposit date: | 2023-03-07 | | Release date: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | Human TRPV1 structure and inhibition by the analgesic SB-366791.
Nat Commun, 14, 2023
|
|
8BCW
 
 | | Photosystem I assembly intermediate of Avena sativa | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Naschberger, A, Amunts, A, Nelson, N. | | Deposit date: | 2022-10-17 | | Release date: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.11 Å) | | Cite: | Photosystem I assembly intermediate of Avena sativa
To Be Published
|
|
