5FN5
 
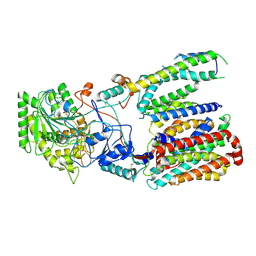 | | Cryo-EM structure of gamma secretase in class 3 of the apo- state ensemble | | Descriptor: | Gamma-secretase subunit APH-1A, Gamma-secretase subunit PEN-2, Nicastrin, ... | | Authors: | Bai, X.C, Rajendra, E, Yang, G.H, Shi, Y.G, Scheres, S.H.W. | | Deposit date: | 2015-11-10 | | Release date: | 2015-12-16 | | Last modified: | 2019-09-11 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Sampling the conformational space of the catalytic subunit of human gamma-secretase.
Elife, 4, 2015
|
|
5FN3
 
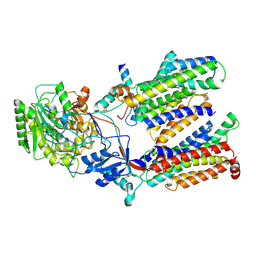 | | Cryo-EM structure of gamma secretase in class 1 of the apo- state ensemble | | Descriptor: | Gamma-secretase subunit APH-1A, Gamma-secretase subunit PEN-2, Nicastrin, ... | | Authors: | Bai, X.C, Rajendra, E, Yang, G.H, Shi, Y.G, Scheres, S.H.W. | | Deposit date: | 2015-11-10 | | Release date: | 2015-12-16 | | Last modified: | 2019-09-11 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Sampling the conformational space of the catalytic subunit of human gamma-secretase.
Elife, 4, 2015
|
|
5FN2
 
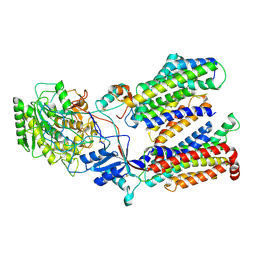 | | Cryo-EM structure of gamma secretase in complex with a drug DAPT | | Descriptor: | Gamma-secretase subunit APH-1A, Gamma-secretase subunit PEN-2, Nicastrin, ... | | Authors: | Bai, X.C, Rajendra, E, Yang, G.H, Shi, Y.G, Scheres, S.H.W. | | Deposit date: | 2015-11-10 | | Release date: | 2015-12-16 | | Last modified: | 2019-09-04 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Sampling the conformational space of the catalytic subunit of human gamma-secretase.
Elife, 4, 2015
|
|
5FN4
 
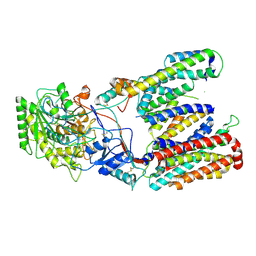 | | Cryo-EM structure of gamma secretase in class 2 of the apo- state ensemble | | Descriptor: | Gamma-secretase subunit APH-1A, Gamma-secretase subunit PEN-2, Nicastrin, ... | | Authors: | Bai, X.C, Rajendra, E, Yang, G.H, Shi, Y.G, Scheres, S.H.W. | | Deposit date: | 2015-11-10 | | Release date: | 2015-12-16 | | Last modified: | 2019-09-11 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Sampling the conformational space of the catalytic subunit of human gamma-secretase.
Elife, 4, 2015
|
|
5GL0
 
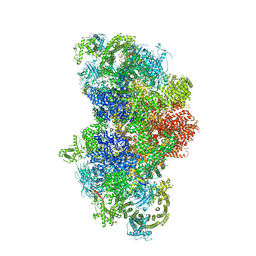 | | Structure of RyR1 in a closed state (C4 conformer) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1A, Ryanodine receptor 1, ZINC ION | | Authors: | Bai, X.C, Yan, Z, Wu, J.P, Yan, N. | | Deposit date: | 2016-07-07 | | Release date: | 2016-08-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The Central domain of RyR1 is the transducer for long-range allosteric gating of channel opening
Cell Res., 26, 2016
|
|
5GKZ
 
 | | Structure of RyR1 in a closed state (C3 conformer) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1A, Ryanodine receptor 1, ZINC ION | | Authors: | Bai, X.C, Yan, Z, Wu, J.P, Yan, N. | | Deposit date: | 2016-07-07 | | Release date: | 2016-08-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The Central domain of RyR1 is the transducer for long-range allosteric gating of channel opening
Cell Res., 26, 2016
|
|
5GL1
 
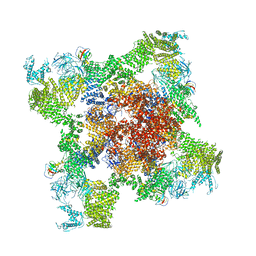 | | Structure of RyR1 in an open state | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1A, Ryanodine receptor 1, ZINC ION | | Authors: | Bai, X.C, Yan, Z, Wu, J.P, Yan, N. | | Deposit date: | 2016-07-07 | | Release date: | 2016-08-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | The Central domain of RyR1 is the transducer for long-range allosteric gating of channel opening
Cell Res., 26, 2016
|
|
5GKY
 
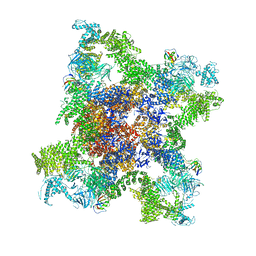 | | Structure of RyR1 in a closed state (C1 conformer) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1A, Ryanodine receptor 1, ZINC ION | | Authors: | Bai, X.C, Yan, Z, Wu, J.P, Yan, N. | | Deposit date: | 2016-07-07 | | Release date: | 2016-08-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The Central domain of RyR1 is the transducer for long-range allosteric gating of channel opening
Cell Res., 26, 2016
|
|
7SL6
 
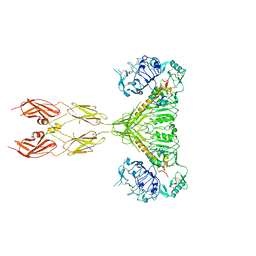 | |
7SL7
 
 | |
7SL2
 
 | |
7SL3
 
 | |
7SL4
 
 | |
7SL1
 
 | |
6XPE
 
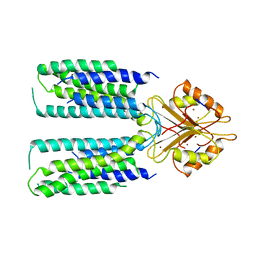 | | Cryo-EM structure of human ZnT8 WT, in the presence of zinc, determined in outward-facing conformation | | Descriptor: | ZINC ION, Zinc transporter 8 | | Authors: | Bai, X.C, Xue, J, Jiang, Y.X. | | Deposit date: | 2020-07-08 | | Release date: | 2020-08-05 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structures of human ZnT8 in both outward- and inward-facing conformations.
Elife, 9, 2020
|
|
6XPF
 
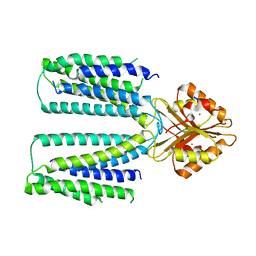 | | Cryo-EM structure of human ZnT8 WT, in the absence of zinc, determined in heterogeneous conformations- one subunit in an inward-facing and the other in an outward-facing conformation | | Descriptor: | ZINC ION, Zinc transporter 8 | | Authors: | Bai, X.C, Xue, J, Jiang, Y.X. | | Deposit date: | 2020-07-08 | | Release date: | 2020-08-05 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Cryo-EM structures of human ZnT8 in both outward- and inward-facing conformations.
Elife, 9, 2020
|
|
6XPD
 
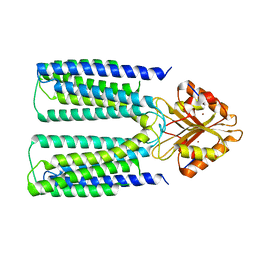 | |
7STI
 
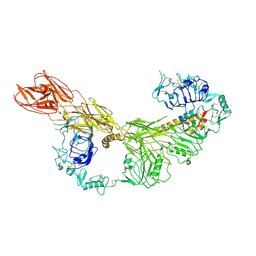 | |
7STJ
 
 | |
7STK
 
 | |
7STH
 
 | |
4V8Z
 
 | | Cryo-EM reconstruction of the 80S-eIF5B-Met-itRNAMet Eukaryotic Translation Initiation Complex | | Descriptor: | 18S RIBOSOMAL RNA, 25S RIBOSOMAL RNA, 40S RIBOSOMAL PROTEIN S0-A, ... | | Authors: | Fernandez, I.S, Bai, X.C, Hussain, T, Kelley, A.C, Lorsch, J.R, Ramakrishnan, V, Scheres, S.H.W. | | Deposit date: | 2013-07-20 | | Release date: | 2014-07-09 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Molecular architecture of a eukaryotic translational initiation complex.
Science, 342, 2013
|
|
4V8Y
 
 | | Cryo-EM reconstruction of the 80S-eIF5B-Met-itRNAMet Eukaryotic Translation Initiation Complex | | Descriptor: | 18S RIBOSOMAL RNA, 25S RIBOSOMAL RNA, 40S RIBOSOMAL PROTEIN S0-A, ... | | Authors: | Fernandez, I.S, Bai, X.C, Hussain, T, Kelley, A.C, Lorsch, J.R, Ramakrishnan, V, Scheres, S.H.W. | | Deposit date: | 2013-07-20 | | Release date: | 2014-07-09 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Molecular architecture of a eukaryotic translational initiation complex.
Science, 342, 2013
|
|
7SII
 
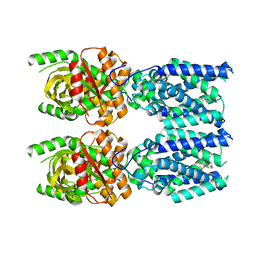 | | Human STING bound to both cGAMP and 1-[(2-chloro-6-fluorophenyl)methyl]-3,3-dimethyl-2-oxo-N-[(2,4,6-trifluorophenyl)methyl]-2,3-dihydro-1H-indole-6-carboxamide (Compound 53) | | Descriptor: | 1-[(2-chloro-6-fluorophenyl)methyl]-3,3-dimethyl-2-oxo-N-[(2,4,6-trifluorophenyl)methyl]-2,3-dihydro-1H-indole-6-carboxamide, Stimulator of interferon genes protein, cGAMP | | Authors: | Lu, D, Shang, G, Jie, L, Lu, Y, Bai, X.C, Zhang, X. | | Deposit date: | 2021-10-14 | | Release date: | 2022-02-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Activation of STING by targeting a pocket in the transmembrane domain.
Nature, 604, 2022
|
|
5GAM
 
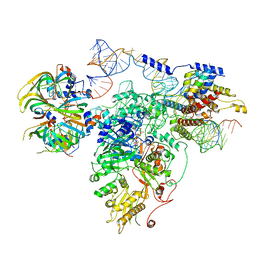 | | Foot region of the yeast spliceosomal U4/U6.U5 tri-snRNP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Pre-mRNA-splicing factor 8, Pre-mRNA-splicing factor SNU114, ... | | Authors: | Nguyen, T.H.D, Galej, W.P, Bai, X.C, Oubridge, C, Scheres, S.H.W, Newman, A.J, Nagai, K. | | Deposit date: | 2015-12-15 | | Release date: | 2016-02-03 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 Angstrom resolution
Nature, 530, 2016
|
|
