1H9P
 
 | | Crystal Structure of Dioclea guianensis Seed Lectin | | Descriptor: | CADMIUM ION, LECTIN ALPHA CHAIN, MANGANESE (II) ION | | Authors: | Romero, A, Wah, D.A, Gallego Del sol, F, Cavada, B.S, Ramos, M.V, Grangeiro, T.B, Sampaio, A.H, Calvete, J.J. | | Deposit date: | 2001-03-16 | | Release date: | 2001-03-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Native and Cd/Cd-Substituted Dioclea Guianensis Seed Lectin. A Novel Manganese-Binding Site and Structural Basis of Dimer-Tetramer Association
J.Mol.Biol., 310, 2001
|
|
1ZYH
 
 | | Structure of a Supercoiling Responsive DNA site | | Descriptor: | 5'-D(*CP*AP*AP*CP*CP*AP*GP*GP*GP*TP*TP*G)-3', 5'-D(*CP*AP*AP*CP*CP*CP*TP*GP*GP*TP*TP*G)-3' | | Authors: | Bae, S.H, Yun, S.H, Sun, D, Lim, H.M, Choi, B.S. | | Deposit date: | 2005-06-10 | | Release date: | 2006-05-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamic basis of a supercoiling-responsive DNA element
Nucleic Acids Res., 34, 2006
|
|
1ZYG
 
 | | Structure of a Supercoiling Responsive DNA Site | | Descriptor: | 5'-D(*CP*AP*AP*CP*CP*CP*GP*GP*GP*TP*TP*G)-3' | | Authors: | Bae, S.H, Yun, S.H, Sun, D, Lim, H.M, Choi, B.S. | | Deposit date: | 2005-06-10 | | Release date: | 2006-05-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamic basis of a supercoiling-responsive DNA element
Nucleic Acids Res., 34, 2006
|
|
5FG5
 
 | | Crystal structure of the bromodomain of human BRPF1 in complex with PFI-4 chemical probe | | Descriptor: | NITRATE ION, Peregrin, ~{N}-(1,3-dimethyl-2-oxidanylidene-6-pyrrolidin-1-yl-benzimidazol-5-yl)-2-methoxy-benzamide | | Authors: | Tallant, C, Owen, D.R, Gerstenberger, B.S, Savitsky, P, Chaikuad, A, Fedorov, O, Nunez-Alonso, G, Filippakopoulos, P, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Muller, S, Brennan, P.E, Knapp, S. | | Deposit date: | 2015-12-20 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the bromodomain of human BRPF1 in complex with PFI-4 chemical probe
To Be Published
|
|
1QZP
 
 | |
1QQV
 
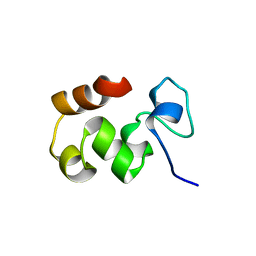 | |
5XF3
 
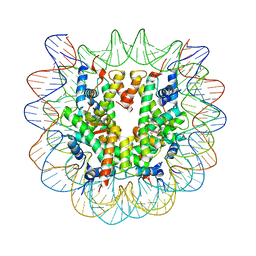 | | Nucleosome core particle with an adduct of a binuclear RAPTA (Ru-arene-phosphaadamantane) compound having a 1,2-diphenylethylenediamine linker (R,R-configuration) | | Descriptor: | (1R,2R)-1,2-diphenylethane-1,2-diamine, DNA (145-MER), Histone H2A type 1-B/E, ... | | Authors: | Ma, Z, Adhireksan, Z, Murray, B.S, Dyson, P.J, Davey, C.A. | | Deposit date: | 2017-04-07 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Nucleosome acidic patch-targeting binuclear ruthenium compounds induce aberrant chromatin condensation
Nat Commun, 8, 2017
|
|
1H4R
 
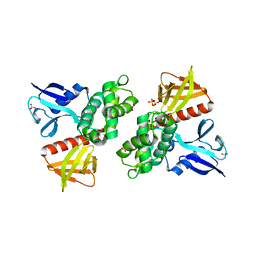 | | Crystal Structure of the FERM domain of Merlin, the Neurofibromatosis 2 Tumor Suppressor Protein. | | Descriptor: | MERLIN, SULFATE ION | | Authors: | Cooper, D.R, Kang, B.S, Sheffield, P, Devedjiev, Y, Derewenda, Z.S. | | Deposit date: | 2001-05-14 | | Release date: | 2002-01-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of the Ferm Domain of Merlin, the Neurofibromatosis Type 2 Gene Product.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
2XVJ
 
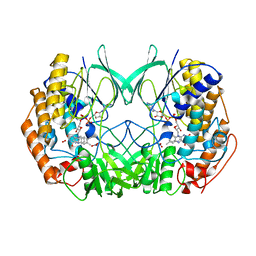 | |
5XF5
 
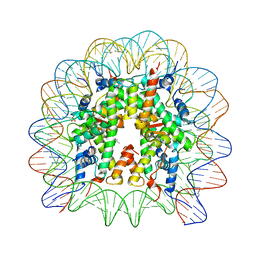 | | Nucleosome core particle with an adduct of a binuclear RAPTA (Ru-arene-phosphaadamantane) compound having a 1,2-diphenylethylenediamine linker (R,S-configuration) | | Descriptor: | (1S,2R)-1,2-diphenylethane-1,2-diamine, DNA (145-MER), Histone H2A type 1-B/E, ... | | Authors: | Ma, Z, Adhireksan, Z, Murray, B.S, Dyson, P.J, Davey, C.A. | | Deposit date: | 2017-04-07 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Nucleosome acidic patch-targeting binuclear ruthenium compounds induce aberrant chromatin condensation
Nat Commun, 8, 2017
|
|
2XVF
 
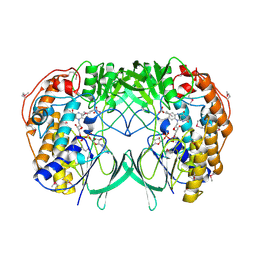 | | Crystal structure of bacterial flavin-containing monooxygenase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FLAVIN-CONTAINING MONOOXYGENASE, GLYCEROL, ... | | Authors: | Cho, H.J, Kang, B.S. | | Deposit date: | 2010-10-26 | | Release date: | 2011-05-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Functional Analysis of Bacterial Flavin-Containing Monooxygenase Reveals its Ping-Pong-Type Reaction Mechanism.
J.Struct.Biol., 175, 2011
|
|
1HR9
 
 | | Yeast Mitochondrial Processing Peptidase beta-E73Q Mutant Complexed with Malate Dehydrogenase Signal Peptide | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MALATE DEHYDROGENASE, MITOCHONDRIAL PROCESSING PEPTIDASE ALPHA SUBUNIT, ... | | Authors: | Taylor, A.B, Smith, B.S, Kitada, S, Kojima, K, Miyaura, H, Otwinowski, Z, Ito, A, Deisenhofer, J. | | Deposit date: | 2000-12-21 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences.
Structure, 9, 2001
|
|
6II2
 
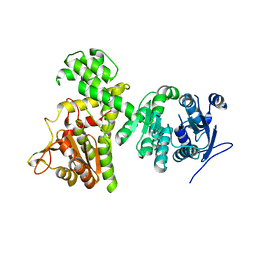 | | Crystal structure of alpha-beta hydrolase (ABH) and Makes Caterpillars Floppy (MCF)-Like effectors of Vibrio vulnificus MO6-24/O | | Descriptor: | Putative RTX-toxin | | Authors: | Lee, Y, Kim, B.S, Choi, S, Lee, E.Y, Park, S, Hwang, J, Kwon, Y, Hyung, J, Lee, C, Eom, S.H, Kim, M.H. | | Deposit date: | 2018-10-03 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Makes caterpillars floppy-like effector-containing MARTX toxins require host ADP-ribosylation factor (ARF) proteins for systemic pathogenicity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6II0
 
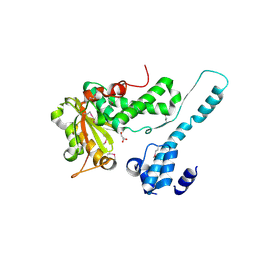 | | Crystal structure of the Makes Caterpillars Floppy (MCF)-Like effector of Vibrio vulnificus MO6-24/O | | Descriptor: | GLYCEROL, Putative RTX-toxin | | Authors: | Lee, Y, Kim, B.S, Choi, S, Lee, E.Y, Park, S, Hwang, J, Kwon, Y, Hyun, J, Lee, C, Eom, S.H, Kim, M.H. | | Deposit date: | 2018-10-03 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Makes caterpillars floppy-like effector-containing MARTX toxins require host ADP-ribosylation factor (ARF) proteins for systemic pathogenicity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6FGM
 
 | | The NMR solution structure of the peptide AC12 from Hypsiboas raniceps | | Descriptor: | ALA-CYS-PHE-LEU-THR-ARG-LEU-GLY-THR-TYR-VAL-CYS | | Authors: | Popov, C.S.F.C, Simas, B.S, Goodfellow, B.J, Bocca, A.L, Andrade, P.B, Pereira, D, Valentao, P, Pereira, P.J.B, Rodrigues, J.E, Veloso Jr, P.H.H, Rezende, T.M.B. | | Deposit date: | 2018-01-11 | | Release date: | 2019-01-09 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Host-defense peptides AC12, DK16 and RC11 with immunomodulatory activity isolated from Hypsiboas raniceps skin secretion.
Peptides, 113, 2019
|
|
6EMH
 
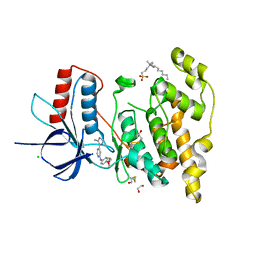 | | Crystal structure of JNK3 in complex with a pyridinylimidazole inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-(4-methyl-1~{H}-imidazol-5-yl)-~{N}-(4-morpholin-4-ylphenyl)pyridin-2-amine, BETA-MERCAPTOETHANOL, ... | | Authors: | Macedo, J.T, Stehle, T, Blaum, B.S. | | Deposit date: | 2017-10-02 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Optimization of a Pyridinylimidazole Scaffold: Shifting the Selectivity from p38 alpha Mitogen-Activated Protein Kinase to c-Jun N-Terminal Kinase 3.
ACS Omega, 3, 2018
|
|
8CXL
 
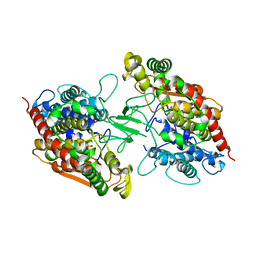 | | Structure of NapH3, a vanadium-dependent haloperoxidase homolog catalyzing the stereospecific alpha-hydroxyketone rearrangement reaction in napyradiomycin biosynthesis | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, NapH3 | | Authors: | Chen, P.Y.-T, Chekan, J.R, Moore, B.S. | | Deposit date: | 2022-05-21 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Basis of Stereospecific Vanadium-Dependent Haloperoxidase Family Enzymes in Napyradiomycin Biosynthesis.
Biochemistry, 61, 2022
|
|
4V4Q
 
 | | Crystal structure of the bacterial ribosome from Escherichia coli at 3.5 A resolution. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Schuwirth, B.S, Borovinskaya, M.A, Hau, C.W, Zhang, W, Vila-Sanjurjo, A, Holton, J.M, Cate, J.H.D. | | Deposit date: | 2005-08-30 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Structures of the bacterial ribosome at 3.5 A resolution.
Science, 310, 2005
|
|
4V4H
 
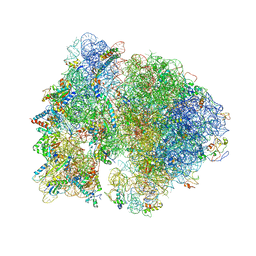 | | Crystal structure of the bacterial ribosome from Escherichia coli in complex with the antibiotic kasugamyin at 3.5A resolution. | | Descriptor: | (1S,2R,3S,4R,5S,6S)-2,3,4,5,6-PENTAHYDROXYCYCLOHEXYL 2-AMINO-4-{[CARBOXY(IMINO)METHYL]AMINO}-2,3,4,6-TETRADEOXY-ALPHA-D-ARABINO-HEXOPYRANOSIDE, 16S RIBOSOMAL RNA, 23S RIBOSOMAL RNA, ... | | Authors: | Schuwirth, B.S, Vila-Sanjurjo, A, Cate, J.H.D. | | Deposit date: | 2006-08-04 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Structural analysis of kasugamycin inhibition of translation.
Nat.Struct.Mol.Biol., 13, 2006
|
|
8FFT
 
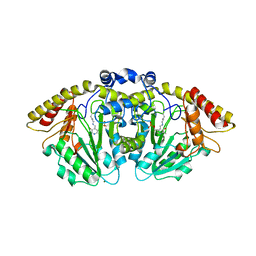 | | Structure of GntC, a PLP-dependent enzyme catalyzing L-enduracididine biosynthesis from (S)-4-hydroxy-L-arginine | | Descriptor: | Aminotransferase class I/II-fold pyridoxal phosphate-dependent enzyme, MAGNESIUM ION | | Authors: | Chen, P.Y.-T, Lima, S.T, Chekan, J.R, Moore, B.S. | | Deposit date: | 2022-12-10 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanistic and Structural Insights into a Divergent PLP-Dependent l-Enduracididine Cyclase from a Toxic Cyanobacterium.
Acs Catalysis, 13, 2023
|
|
8FFU
 
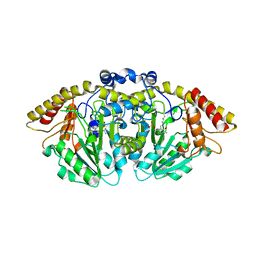 | | Structure of GntC, a PLP-dependent enzyme catalyzing L-enduracididine biosynthesis from (S)-4-hydroxy-L-arginine, with the substrate bound | | Descriptor: | (2S,4S)-5-carbamimidamido-4-hydroxy-2-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]pentanoic acid (non-preferred name), Aminotransferase class I/II-fold pyridoxal phosphate-dependent enzyme, MAGNESIUM ION | | Authors: | Chen, P.Y.-T, Moore, B.S. | | Deposit date: | 2022-12-10 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Mechanistic and Structural Insights into a Divergent PLP-Dependent l-Enduracididine Cyclase from a Toxic Cyanobacterium.
Acs Catalysis, 13, 2023
|
|
4V4G
 
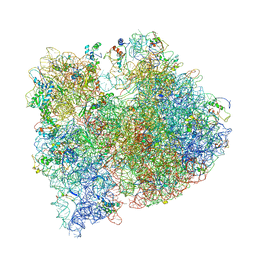 | | Crystal structure of five 70s ribosomes from Escherichia Coli in complex with protein Y. | | Descriptor: | 16S ribosomal RNA, 23S RIBOSOMAL RNA, 30S ribosomal protein S10, ... | | Authors: | Vila-Sanjurjo, A, Schuwirth, B.S, Hau, C.W, Cate, J.H. | | Deposit date: | 2004-10-06 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (11.5 Å) | | Cite: | Structural basis for the control of translation initiation during stress.
Nat.Struct.Mol.Biol., 11, 2004
|
|
6VL1
 
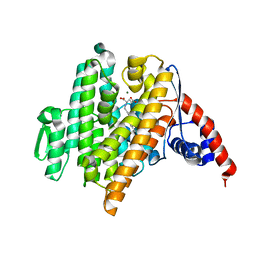 | | Crystal Structure of the N-prenyltransferase DabA in Complex with NGG and Mg2+ | | Descriptor: | DabA, MAGNESIUM ION, N-[(2E)-3,7-dimethylocta-2,6-dien-1-yl]-L-glutamic acid | | Authors: | Chekan, J.R, Noel, J.P, Moore, B.S. | | Deposit date: | 2020-01-22 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Algal neurotoxin biosynthesis repurposes the terpene cyclase structural fold into anN-prenyltransferase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VKZ
 
 | |
2KYD
 
 | | RDC and RCSA refinement of an A-form RNA: Improvements in Major Groove Width | | Descriptor: | RNA (5'-R(*CP*UP*AP*GP*UP*UP*AP*GP*CP*UP*AP*AP*CP*UP*AP*G)-3') | | Authors: | Tolbert, B.S, Summers, M.F, Miyazaki, Y, Barton, S, Kinde, B, Stark, P, Singh, R, Bax, A, Case, D. | | Deposit date: | 2010-05-24 | | Release date: | 2010-07-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Major groove width variations in RNA structures determined by NMR and impact of 13C residual chemical shift anisotropy and 1H-13C residual dipolar coupling on refinement.
J.Biomol.Nmr, 47, 2010
|
|
