5FGT
 
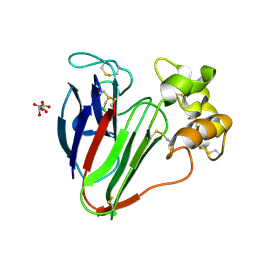 | | Thaumatin solved by native sulphur-SAD using free-electron laser radiation | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Nass, K.J, Meinhart, A, Barends, T.R.M, Foucar, L, Gorel, A, Aquila, A, Botha, S, Doak, R.B, Koglin, J, Liang, M, Shoeman, R.L, Williams, G.K, Boutet, S, Schlichting, I. | | Deposit date: | 2015-12-21 | | Release date: | 2016-06-08 | | Last modified: | 2018-11-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Protein structure determination by single-wavelength anomalous diffraction phasing of X-ray free-electron laser data.
Iucrj, 3, 2016
|
|
5FGX
 
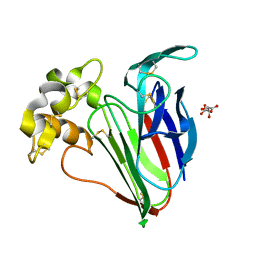 | | Thaumatin solved by native sulphur SAD using synchrotron radiation | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Nass, K.J, Meinhart, A, Barends, T.R.M, Foucar, L, Gorel, A, Aquila, A, Botha, S, Doak, R.B, Koglin, J, Liang, M, Shoeman, R.L, Williams, G.J, Boutet, S, Schlichting, I. | | Deposit date: | 2015-12-21 | | Release date: | 2016-06-08 | | Method: | X-RAY DIFFRACTION (2.134 Å) | | Cite: | Protein structure determination by single-wavelength anomalous diffraction phasing of X-ray free-electron laser data.
Iucrj, 3, 2016
|
|
6WQA
 
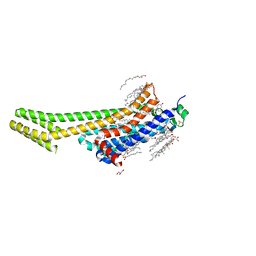 | | 2.0A angstrom A2a adenosine receptor structure using XFEL data collected in helium atmosphere. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Lee, M.-Y, Geiger, J, Ishchenko, A, Han, G.W, Barty, A, White, T.A, Gati, C, Batyuk, A, Hunter, M.S, Aquila, A, Boutet, S, Weierstall, U, Cherezov, V, Liu, W. | | Deposit date: | 2020-04-28 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Harnessing the power of an X-ray laser for serial crystallography of membrane proteins crystallized in lipidic cubic phase
Iucrj, 7, 2020
|
|
3PCQ
 
 | | Femtosecond X-ray protein Nanocrystallography | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Chapman, H.N, Fromme, P, Barty, A, White, T.A, Kirian, R.A, Aquila, A, Hunter, M.S, Schulz, J, Deponte, D.P, Weierstall, U, Doak, R.B, Maia, F.R.N.C, Martin, A.V, Schlichting, I, Lomb, L, Coppola, N, Shoeman, R.L, Epp, S.W, Hartmann, R, Rolles, D, Rudenko, A, Foucar, L, Kimmel, N, Weidenspointner, G, Holl, P, Liang, M, Barthelmess, M, Caleman, C, Boutet, S, Bogan, M.J, Krzywinski, J, Bostedt, C, Bajt, S, Gumprecht, L, Rudek, B, Erk, B, Schmidt, C, Homke, A, Reich, C, Pietschner, D, Struder, L, Hauser, G, Gorke, H, Ullrich, J, Herrmann, S, Schaller, G, Schopper, F, Soltau, H, Kuhnel, K.-U, Messerschmidt, M, Bozek, J.D, Hau-Riege, S.P, Frank, M, Hampton, C.Y, Sierra, R, Starodub, D, Williams, G.J, Hajdu, J, Timneanu, N, Seibert, M.M, Andreasson, J, Rocker, A, Jonsson, O, Svenda, M, Stern, S, Nass, K, Andritschke, R, Schroter, C.-D, Krasniqi, F, Bott, M, Schmidt, K.E, Wang, X, Grotjohann, I, Holton, J.M, Barends, T.R.M, Neutze, R, Marchesini, S, Fromme, R, Schorb, S, Rupp, D, Adolph, M, Gorkhover, T, Andersson, I, Hirsemann, H, Potdevin, G, Graafsma, H, Nilsson, B, Spence, J.C.H. | | Deposit date: | 2010-10-21 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (8.984 Å) | | Cite: | Femtosecond X-ray protein nanocrystallography.
Nature, 470, 2011
|
|
5BR8
 
 | | Ambient-temperature crystal structure of 30S ribosomal subunit from Thermus thermophilus in complex with paromomycin | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Sierra, R.G, Gati, C, Laksmono, H, Dao, E.H, Gul, S, Fuller, F, Kern, J, Chatterjee, R, Ibrahim, M, Brewster, A, Young, I.D, Michels-Clark, T, Aquila, A, Mengning, L, Hunter, M.S, Koglin, J.E, Boutet, S, Junco, E.A, Hayes, B, Bogan, M.J, Hampton, C.Y, Puglisi, E.V, Sauter, N.K, Stan, C.A, Zouni, A, Yano, J, Yachandra, V.K, Soltis, S.M, Puglisi, J.D, DeMirci, H. | | Deposit date: | 2015-05-29 | | Release date: | 2015-11-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Ambient-temperature crystal structure of 30S ribosomal subunit from Thermus thermophilus in complex with paromomycin
To Be Published
|
|
7L6W
 
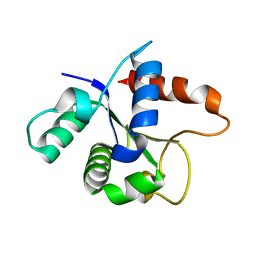 | | SFX structure of the MyD88 TIR domain higher-order assembly | | Descriptor: | Myeloid differentiation primary response protein MyD88 | | Authors: | Clabbers, M.T.B, Holmes, S, Muusse, T.W, Vajjhala, P, Thygesen, S.J, Malde, A.K, Hunter, D.J.B, Croll, T.I, Flueckiger, L, Nanson, J.D, Rahaman, M.H, Aquila, A, Hunter, M.S, Liang, M, Yoon, C.H, Zhao, J, Zatsepin, N.A, Abbey, B, Sierecki, E, Gambin, Y, Stacey, K.J, Darmanin, C, Kobe, B, Xu, H, Ve, T. | | Deposit date: | 2020-12-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | MyD88 TIR domain higher-order assembly interactions revealed by microcrystal electron diffraction and serial femtosecond crystallography.
Nat Commun, 12, 2021
|
|
5TIS
 
 | | Room temperature XFEL structure of the native, doubly-illuminated photosystem II complex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Fuller, F, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Hussein, R, Zhang, M, Douthit, L, Kubin, M, de Lichtenberg, C, Pham, L.V, Nilsson, H, Cheah, M.H, Shevela, D, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Pastor, E, Weninger, C, Fransson, T, Lassalle, L, Braeuer, P, Aller, P, Docker, P.T, Andi, B, Orville, A.M, Glownia, J.M, Nelson, S, Sikorski, M, Zhu, D, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Watermann, D.G, Evans, G, Wernet, P, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-10-03 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25000381 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
4AC5
 
 | | Lipidic sponge phase crystal structure of the Bl. viridis reaction centre solved using serial femtosecond crystallography | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Johansson, L.C, Arnlund, D, White, T.A, Katona, G, DePonte, D.P, Weierstall, U, Doak, R.B, Shoeman, R.L, Lomb, L, Malmerberg, E, Davidsson, J, Nass, K, Liang, M, Andreasson, J, Aquila, A, Bajt, S, Barthelmess, M, Barty, A, Bogan, M.J, Bostedt, C, Bozek, J.D, Caleman, C, Coffee, R, Coppola, N, Ekeberg, T, Epp, S.W, Erk, B, Fleckenstein, H, Foucar, L, Graafsma, H, Gumprecht, L, Hajdu, J, Hampton, C.Y, Hartmann, R, Hartmann, A, Hauser, G, Hirsemann, H, Holl, P, Hunter, M.S, Kassemeyer, S, Kimmel, N, Kirian, R.A, Maia, F.R.N.C, Marchesini, S, Martin, A.V, Reich, C, Rolles, D, Rudek, B, Rudenko, A, Schlichting, I, Schulz, J, Seibert, M.M, Sierra, R, Soltau, H, Starodub, D, Stellato, F, Stern, S, Struder, L, Timneanu, N, Ullrich, J, Wahlgren, W.Y, Wang, X, Weidenspointner, G, Wunderer, C, Fromme, P, Chapman, H.N, Spence, J.C.H, Neutze, R. | | Deposit date: | 2011-12-14 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (8.2 Å) | | Cite: | Lipidic Phase Membrane Protein Serial Femtosecond Crystallography.
Nat.Methods, 9, 2012
|
|
4CAS
 
 | | Serial femtosecond crystallography structure of a photosynthetic reaction center | | Descriptor: | (2E,6E,10E,14E,18E,22E,26E)-3,7,11,15,19,23,27,31-OCTAMETHYLDOTRIACONTA-2,6,10,14,18,22,26,30-OCTAENYL TRIHYDROGEN DIPHOSPHATE, 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL A, ... | | Authors: | Johansson, L.C, Arnlund, D, Katona, G, White, T.A, Barty, A, DePonte, D.P, Shoeman, R.L, Wickstrand, C, Sharma, A, Williams, G.J, Aquila, A, Bogan, M.J, Caleman, C, Davidsson, J, Doak, R.B, Frank, M, Fromme, R, Galli, L, Grotjohann, I, Hunter, M.S, Kassemeyer, S, Kirian, R.A, Kupitz, C, Liang, M, Lomb, L, Malmerberg, E, Martin, A.V, Messerschmidt, M, Nass, K, Redecke, L, Seibert, M.M, Sjohamn, J, Steinbrener, J, Stellato, F, Wang, D, Wahlgren, W.Y, Weierstall, U, Westenhoff, S, Zatsepin, N.A, Boutet, S, Spence, J.C.H, Schlichting, I, Chapman, H.N, Fromme, P, Neutze, R. | | Deposit date: | 2013-10-09 | | Release date: | 2013-12-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of a photosynthetic reaction centre determined by serial femtosecond crystallography.
Nat Commun, 4, 2013
|
|
4HWY
 
 | | Trypanosoma brucei procathepsin B solved from 40 fs free-electron laser pulse data by serial femtosecond X-ray crystallography | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cysteine peptidase C (CPC), beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Redecke, L, Nass, K, DePonte, D.P, White, T.A, Rehders, D, Barty, A, Stellato, F, Liang, M, Barends, T.R.M, Boutet, S, Williams, G.W, Messerschmidt, M, Seibert, M.M, Aquila, A, Arnlund, D, Bajt, S, Barth, T, Bogan, M.J, Caleman, C, Chao, T.-C, Doak, R.B, Fleckenstein, H, Frank, M, Fromme, R, Galli, L, Grotjohann, I, Hunter, M.S, Johansson, L.C, Kassemeyer, S, Katona, G, Kirian, R.A, Koopmann, R, Kupitz, C, Lomb, L, Martin, A.V, Mogk, S, Neutze, R, Shoemann, R.L, Steinbrener, J, Timneanu, N, Wang, D, Weierstall, U, Zatsepin, N.A, Spence, J.C.H, Fromme, P, Schlichting, I, Duszenko, M, Betzel, C, Chapman, H. | | Deposit date: | 2012-11-09 | | Release date: | 2012-12-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Natively inhibited Trypanosoma brucei cathepsin B structure determined by using an X-ray laser.
Science, 339, 2013
|
|
5K2D
 
 | | 1.9A angstrom A2a adenosine receptor structure with MR phasing using XFEL data | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Batyuk, A, Galli, L, Ishchenko, A, Han, G.W, Gati, C, Popov, P, Lee, M.-Y, Stauch, B, White, T.A, Barty, A, Aquila, A, Hunter, M.S, Liang, M, Boutet, S, Pu, M, Liu, Z.-J, Nelson, G, James, D, Li, C, Zhao, Y, Spence, J.C.H, Liu, W, Fromme, P, Katritch, V, Weierstall, U, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Native phasing of x-ray free-electron laser data for a G protein-coupled receptor.
Sci Adv, 2, 2016
|
|
5K2B
 
 | | 2.5 angstrom A2a adenosine receptor structure with MR phasing using XFEL data | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Batyuk, A, Galli, L, Ishchenko, A, Han, G.W, Gati, C, Popov, P, Lee, M.-Y, Stauch, B, White, T.A, Barty, A, Aquila, A, Hunter, M.S, Liang, M, Boutet, S, Pu, M, Liu, Z.-J, Nelson, G, James, D, Li, C, Zhao, Y, Spence, J.C.H, Liu, W, Fromme, P, Katritch, V, Weierstall, U, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Native phasing of x-ray free-electron laser data for a G protein-coupled receptor.
Sci Adv, 2, 2016
|
|
5K2C
 
 | | 1.9 angstrom A2a adenosine receptor structure with sulfur SAD phasing and phase extension using XFEL data | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a/Soluble cytochrome b562 chimera, ... | | Authors: | Batyuk, A, Galli, L, Ishchenko, A, Han, G.W, Gati, C, Popov, P, Lee, M.-Y, Stauch, B, White, T.A, Barty, A, Aquila, A, Hunter, M.S, Liang, M, Boutet, S, Pu, M, Liu, Z.-J, Nelson, G, James, D, Li, C, Zhao, Y, Spence, J.C.H, Liu, W, Fromme, P, Katritch, V, Weierstall, U, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-21 | | Last modified: | 2018-11-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Native phasing of x-ray free-electron laser data for a G protein-coupled receptor.
Sci Adv, 2, 2016
|
|
5K2A
 
 | | 2.5 angstrom A2a adenosine receptor structure with sulfur SAD phasing using XFEL data | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a/Soluble cytochrome b562 chimera, ... | | Authors: | Batyuk, A, Galli, L, Ishchenko, A, Han, G.W, Gati, C, Popov, P, Lee, M.-Y, Stauch, B, White, T.A, Barty, A, Aquila, A, Hunter, M.S, Liang, M, Boutet, S, Pu, M, Liu, Z.-J, Nelson, G, James, D, Li, C, Zhao, Y, Spence, J.C.H, Liu, W, Fromme, P, Katritch, V, Weierstall, U, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-21 | | Last modified: | 2018-11-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Native phasing of x-ray free-electron laser data for a G protein-coupled receptor.
Sci Adv, 2, 2016
|
|
7BER
 
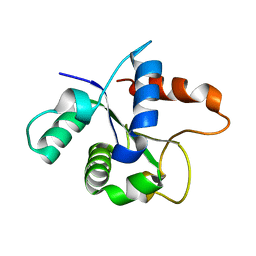 | | SFX structure of the MyD88 TIR domain higher-order assembly (solved, rebuilt and refined using an identical protocol to the MicroED structure of the MyD88 TIR domain higher-order assembly) | | Descriptor: | Myeloid differentiation primary response protein MyD88 | | Authors: | Clabbers, M.T.B, Holmes, S, Muusse, T.W, Vajjhala, P, Thygesen, S.J, Malde, A.K, Hunter, D.J.B, Croll, T.I, Nanson, J.D, Rahaman, M.H, Aquila, A, Hunter, M.S, Liang, M, Yoon, C.H, Zhao, J, Zatsepin, N.A, Abbey, B, Sierecki, E, Gambin, Y, Darmanin, C, Kobe, B, Xu, H, Ve, T. | | Deposit date: | 2020-12-24 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | MyD88 TIR domain higher-order assembly interactions revealed by microcrystal electron diffraction and serial femtosecond crystallography.
Nat Commun, 12, 2021
|
|
7BEQ
 
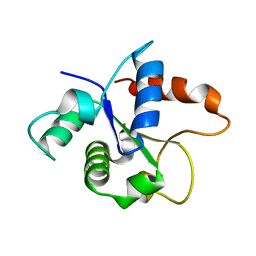 | | MicroED structure of the MyD88 TIR domain higher-order assembly | | Descriptor: | Myeloid differentiation primary response protein MyD88 | | Authors: | Clabbers, M.T.B, Holmes, S, Muusse, T.W, Vajjhala, P, Thygesen, S.J, Malde, A.K, Hunter, D.J.B, Croll, T.I, Nanson, J.D, Rahaman, M.H, Aquila, A, Hunter, M.S, Liang, M, Yoon, C.H, Zhao, J, Zatsepin, N.A, Abbey, B, Sierecki, E, Gambin, Y, Darmanin, C, Kobe, B, Xu, H, Ve, T. | | Deposit date: | 2020-12-24 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON CRYSTALLOGRAPHY (3 Å) | | Cite: | MyD88 TIR domain higher-order assembly interactions revealed by microcrystal electron diffraction and serial femtosecond crystallography.
Nat Commun, 12, 2021
|
|
5KAI
 
 | | NH3-bound RT XFEL structure of Photosystem II 500 ms after the 2nd illumination (2F) at 2.8 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Fuller, F, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Waterman, D.G, Evans, G, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-06-01 | | Release date: | 2016-11-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.80000925 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
5KAF
 
 | | RT XFEL structure of Photosystem II in the dark state at 3.0 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Fuller, F, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Waterman, D.G, Evans, G, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-06-01 | | Release date: | 2016-11-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.00001 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
4ETC
 
 | | Lysozyme, room temperature, 24 kGy dose | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.906 Å) | | Cite: | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
4ETB
 
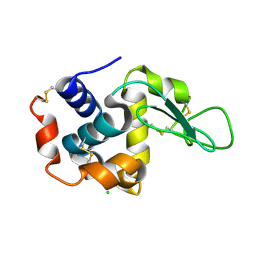 | | lysozyme, room temperature, 200 kGy dose | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.908 Å) | | Cite: | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
4ET8
 
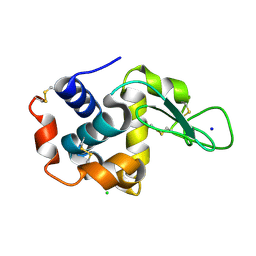 | | Hen egg-white lysozyme solved from 40 fs free-electron laser pulse data | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
4ETE
 
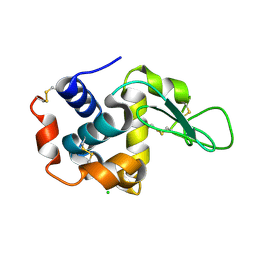 | | Lysozyme, room-temperature, rotating anode, 0.0021 MGy | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
4ETA
 
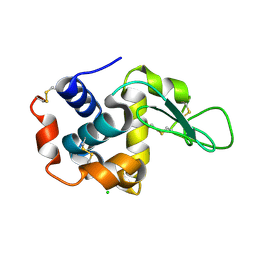 | | Lysozyme, room temperature, 400 kGy dose | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
4ET9
 
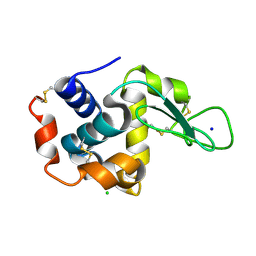 | | Hen egg-white lysozyme solved from 5 fs free-electron laser pulse data | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
4ETD
 
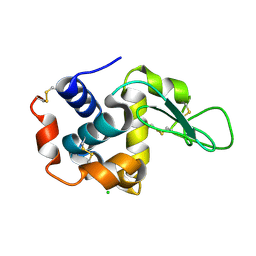 | | Lysozyme, room-temperature, rotating anode, 0.0026 MGy | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
