2PAC
 
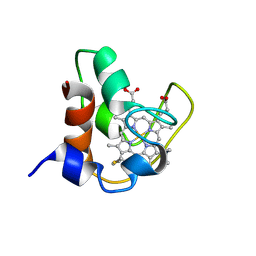 | | SOLUTION STRUCTURE OF FE(II) CYTOCHROME C551 FROM PSEUDOMONAS AERUGINOSA AS DETERMINED BY TWO-DIMENSIONAL 1H NMR | | Descriptor: | CYTOCHROME C551, HEME C | | Authors: | Detlefsen, D.J, Thanabal, V, Pecoraro, V.L, Wagner, G. | | Deposit date: | 1993-05-05 | | Release date: | 1993-10-31 | | Last modified: | 2021-03-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Fe(II) cytochrome c551 from Pseudomonas aeruginosa as determined by two-dimensional 1H NMR.
Biochemistry, 30, 1991
|
|
1AGG
 
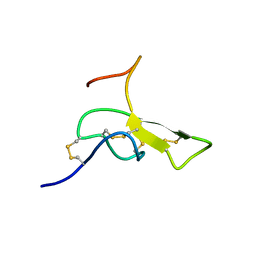 | | THE SOLUTION STRUCTURE OF OMEGA-AGA-IVB, A P-TYPE CALCIUM CHANNEL ANTAGONIST FROM THE VENOM OF AGELENOPSIS APERTA | | Descriptor: | OMEGA-AGATOXIN-IVB | | Authors: | Reily, M.D, Thanabal, V, Adams, M.E. | | Deposit date: | 1995-11-03 | | Release date: | 1996-03-08 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | The solution structure of omega-Aga-IVB, a P-type calcium channel antagonist from venom of the funnel web spider, Agelenopsis aperta.
J.Biomol.NMR, 5, 1995
|
|
1GHU
 
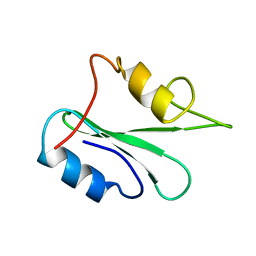 | | NMR solution structure of growth factor receptor-bound protein 2 (GRB2) SH2 domain, 24 structures | | Descriptor: | GRB2 | | Authors: | Thornton, K.H, Mueller, W.T, Mcconnell, P, Zhu, G, Saltiel, A.R, Thanabal, V. | | Deposit date: | 1996-08-05 | | Release date: | 1997-01-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance solution structure of the growth factor receptor-bound protein 2 Src homology 2 domain.
Biochemistry, 35, 1996
|
|
6MNY
 
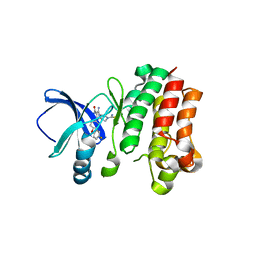 | |
1EIT
 
 | | NMR STUDY OF MU-AGATOXIN | | Descriptor: | MU-AGATOXIN-I | | Authors: | Omecinsky, D.O, Reily, M.D. | | Deposit date: | 1995-12-14 | | Release date: | 1996-04-03 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure analysis of mu-agatoxins: further evidence for common motifs among neurotoxins with diverse ion channel specificities.
Biochemistry, 35, 1996
|
|
1AW6
 
 | | GAL4 (CD), NMR, 24 STRUCTURES | | Descriptor: | CADMIUM ION, GAL4 (CD) | | Authors: | Baleja, J.D, Wagner, G. | | Deposit date: | 1997-10-10 | | Release date: | 1998-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of the DNA-binding domain of GAL4 and use of 3J(113Cd,1H) in structure determination.
J.Biomol.NMR, 10, 1997
|
|
1IVA
 
 | |
2W6Z
 
 | |
2W6O
 
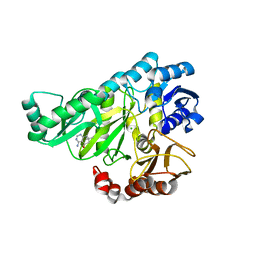 | | Crystal structure of Biotin carboxylase from E. coli in complex with 4-Amino-7,7-dimethyl-7,8-dihydro-quinazolinone fragment | | Descriptor: | 4-amino-7,7-dimethyl-7,8-dihydroquinazolin-5(6H)-one, BIOTIN CARBOXYLASE, CHLORIDE ION | | Authors: | Mochalkin, I, Miller, J.R. | | Deposit date: | 2008-12-18 | | Release date: | 2009-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of Antibacterial Biotin Carboxylase Inhibitors by Virtual Screening and Fragment-Based Approaches.
Acs Chem.Biol., 4, 2009
|
|
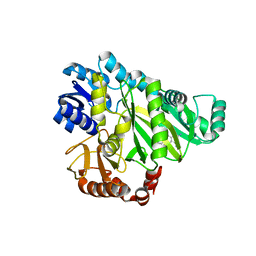
2W6P
 
 | |
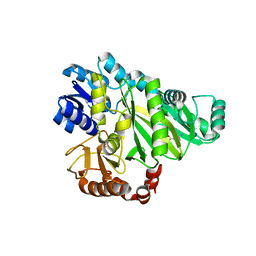
2W6M
 
 | |
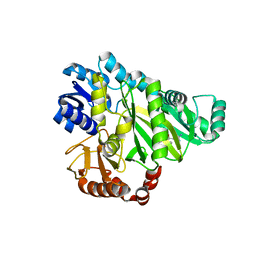
2W6N
 
 | |
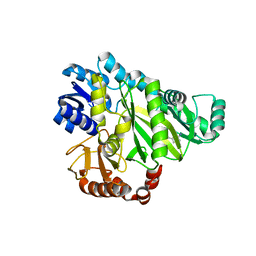
2W70
 
 | |
2W6Q
 
 | | Crystal structure of Biotin carboxylase from E. coli in complex with the triazine-2,4-diamine fragment | | Descriptor: | 6-(2-phenoxyethoxy)-1,3,5-triazine-2,4-diamine, BIOTIN CARBOXYLASE, CHLORIDE ION | | Authors: | Mochalkin, I, Miller, J.R. | | Deposit date: | 2008-12-18 | | Release date: | 2009-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of Antibacterial Biotin Carboxylase Inhibitors by Virtual Screening and Fragment-Based Approaches.
Acs Chem.Biol., 4, 2009
|
|
2W71
 
 | |
