1LVA
 
 | |
1DD5
 
 | | CRYSTAL STRUCTURE OF THERMOTOGA MARITIMA RIBOSOME RECYCLING FACTOR, RRF | | Descriptor: | ACETIC ACID, RIBOSOME RECYCLING FACTOR | | Authors: | Selmer, M, Al-Karadaghi, S, Hirokawa, G, Kaji, A, Liljas, A. | | Deposit date: | 1999-11-08 | | Release date: | 1999-12-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of Thermotoga maritima ribosome recycling factor: a tRNA mimic.
Science, 286, 1999
|
|
4V51
 
 | | Structure of the Thermus thermophilus 70S ribosome complexed with mRNA, tRNA and paromomycin | | Descriptor: | 16S ribosomal RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Selmer, M, Dunham, C.M, Murphy, F.V, Weixlbaumer, A, Petry, S, Weir, J.R, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2006-07-31 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the 70S ribosome complexed with mRNA and tRNA.
Science, 313, 2006
|
|
6FZB
 
 | | AadA in complex with ATP, magnesium and streptomycin | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kanchugal P, S, Selmer, M. | | Deposit date: | 2018-03-14 | | Release date: | 2018-06-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural mechanism of AadA, a dual-specificity aminoglycoside adenylyltransferase fromSalmonella enterica.
J. Biol. Chem., 293, 2018
|
|
4CS6
 
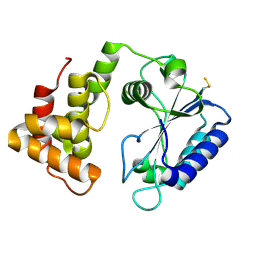 | | Crystal structure of AadA - an aminoglycoside adenyltransferase | | Descriptor: | AMINOGLYCOSIDE ADENYLTRANSFERASE | | Authors: | Chen, Y, Nasvall, J, Andersson, D.I, Selmer, M. | | Deposit date: | 2014-03-05 | | Release date: | 2015-03-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structure of Aada from Salmonella Enterica: A Monomeric Aminoglycoside (3'')(9) Adenyltransferase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8BB1
 
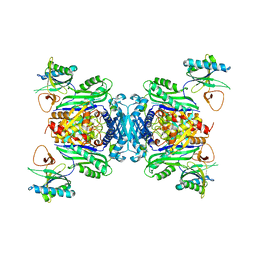 | |
8P18
 
 | | E167K RF2 on E. coli 70S release complex with UGG (Structure III) | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Pundir, S, Larsson, D.S.D, Selmer, M, Sanyal, S. | | Deposit date: | 2023-05-11 | | Release date: | 2024-05-22 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | The compensatory mechanism of a naturally evolved
E167K RF2 counteracting the loss of
RF1 in bacteria
To Be Published
|
|
4V4T
 
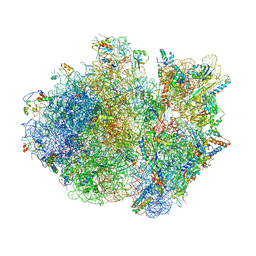 | | Crystal structure of the whole ribosomal complex with a stop codon in the A-site. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Petry, S, Brodersen, D.E, Murphy IV, F.V, Dunham, C.M, Selmer, M, Tarry, M.J, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2005-10-12 | | Release date: | 2014-07-09 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (6.46 Å) | | Cite: | Crystal Structures of the Ribosome in Complex with Release Factors RF1 and RF2 Bound to a Cognate Stop Codon.
Cell(Cambridge,Mass.), 123, 2005
|
|
7ODE
 
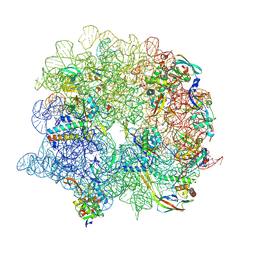 | | E. coli 50S ribosome LiCl core particle | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Larsson, D.S.D, Selmer, M. | | Deposit date: | 2021-04-29 | | Release date: | 2022-06-01 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structural Consequences of Deproteinating the 50S Ribosome.
Biomolecules, 12, 2022
|
|
6NVM
 
 | |
5LUH
 
 | | AadA E87Q in complex with ATP, calcium and streptomycin | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Stern, A.L, Van der Verren, S.E, Selmer, M. | | Deposit date: | 2016-09-08 | | Release date: | 2018-01-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural mechanism of AadA, a dual-specificity aminoglycoside adenylyltransferase fromSalmonella enterica.
J.Biol.Chem., 293, 2018
|
|
5LPA
 
 | | AadA E87Q in complex with ATP, calcium and dihydrostreptomycin | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Stern, A.L, Van der Verren, S.E, Selmer, M. | | Deposit date: | 2016-08-12 | | Release date: | 2018-01-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural mechanism of AadA, a dual-specificity aminoglycoside adenylyltransferase fromSalmonella enterica.
J.Biol.Chem., 293, 2018
|
|
6SXJ
 
 | |
4V5A
 
 | | Structure of the Ribosome Recycling Factor bound to the Thermus thermophilus 70S ribosome with mRNA, ASL-Phe and tRNA-fMet | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Weixlbaumer, A, Petry, S, Dunham, C.M, Selmer, M, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2007-06-28 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the ribosome recycling factor bound to the ribosome.
Nat. Struct. Mol. Biol., 14, 2007
|
|
1MZP
 
 | | Structure of the L1 protuberance in the ribosome | | Descriptor: | 50s ribosomal protein L1P, MAGNESIUM ION, fragment of 23S rRNA | | Authors: | Nikulin, A, Eliseikina, I, Tishchenko, S, Nevskaya, N, Davydova, N, Platonova, O, Piendl, W, Selmer, M, Liljas, A, Zimmermann, R, Garber, M, Nikonov, S. | | Deposit date: | 2002-10-09 | | Release date: | 2003-01-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of the L1 protuberance in the ribosome.
Nat.Struct.Biol., 10, 2003
|
|
1FEU
 
 | | CRYSTAL STRUCTURE OF RIBOSOMAL PROTEIN TL5, ONE OF THE CTC FAMILY PROTEINS, COMPLEXED WITH A FRAGMENT OF 5S RRNA. | | Descriptor: | 19 NT FRAGMENT OF 5S RRNA, 21 NT FRAGMENT OF 5S RRNA, 50S RIBOSOMAL PROTEIN L25, ... | | Authors: | Fedorov, R.V, Meshcheryakov, V.A, Gongadze, G.M, Fomenkova, N.P, Nevskaya, N.A, Selmer, M, Laurberg, M, Kristensen, O, Al-Karadaghi, S, Liljas, A, Garber, M.B, Nikonov, S.V. | | Deposit date: | 2000-07-23 | | Release date: | 2001-06-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of ribosomal protein TL5 complexed with RNA provides new insights into the CTC family of stress proteins.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
7KB0
 
 | | O-acety-L-homoserine aminocarboxypropyltransferase (MetY) from Thermotoga maritima with pyridoxal-5-phosphate (PLP) bound in the internal aldimine state | | Descriptor: | O-acetyl-L-homoserine sulfhydrylase | | Authors: | Brewster, J.L, Pachl, P, Squire, C, Selmer, M, Patrick, W.M. | | Deposit date: | 2020-10-01 | | Release date: | 2021-06-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures and kinetics of Thermotoga maritima MetY reveal new insights into the predominant sulfurylation enzyme of bacterial methionine biosynthesis.
J.Biol.Chem., 296, 2021
|
|
7KB1
 
 | | Complex of O-acety-L-homoserine aminocarboxypropyltransferase (MetY) from Thermotoga maritima and a key reaction intermediate | | Descriptor: | (2E)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]but-3-enoic acid, MAGNESIUM ION, O-acetyl-L-homoserine sulfhydrylase, ... | | Authors: | Brewster, J.L, Pachl, P, Squire, C, Selmer, M, Patrick, W.M. | | Deposit date: | 2020-10-01 | | Release date: | 2021-06-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures and kinetics of Thermotoga maritima MetY reveal new insights into the predominant sulfurylation enzyme of bacterial methionine biosynthesis.
J.Biol.Chem., 296, 2021
|
|
4V5F
 
 | | The structure of the ribosome with elongation factor G trapped in the post-translocational state | | Descriptor: | 16S ribosomal RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Gao, Y.-G, Selmer, M, Dunham, C.M, Weixlbaumer, A, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2009-09-01 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The structure of the ribosome with elongation factor G trapped in the posttranslocational state.
Science, 326, 2009
|
|
4V4R
 
 | | Crystal structure of the whole ribosomal complex. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Petry, S, Brodersen, D.E, Murphy IV, F.V, Dunham, C.M, Selmer, M, Tarry, M.J, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2005-09-30 | | Release date: | 2014-07-09 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (5.9 Å) | | Cite: | Crystal Structures of the Ribosome in Complex with Release Factors RF1 and RF2 Bound to a Cognate Stop Codon.
Cell(Cambridge,Mass.), 123, 2005
|
|
4V4S
 
 | | Crystal structure of the whole ribosomal complex. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Petry, S, Brodersen, D.E, Murphy IV, F.V, Dunham, C.M, Selmer, M, Tarry, M.J, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2005-10-12 | | Release date: | 2014-07-09 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (6.76 Å) | | Cite: | Crystal Structures of the Ribosome in Complex with Release Factors RF1 and RF2 Bound to a Cognate Stop Codon.
Cell(Cambridge,Mass.), 123, 2005
|
|
6ZM9
 
 | | PHAGE SAM LYASE IN COMPLEX WITH S-METHYL-5'-THIOADENOSINE | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, Chains: A, PHOSPHATE ION, ... | | Authors: | Guo, X, Kanchugal P, S, Selmer, M. | | Deposit date: | 2020-07-02 | | Release date: | 2021-02-24 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and mechanism of a phage-encoded SAM lyase revises catalytic function of enzyme family.
Elife, 10, 2021
|
|
6ZMG
 
 | |
6ZNB
 
 | | PHAGE SAM LYASE IN APO STATE | | Descriptor: | PHOSPHATE ION, Phage SAM lyase Svi3-3 | | Authors: | Eckhard, U, Kanchugal P, S, Selmer, M. | | Deposit date: | 2020-07-06 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and mechanism of a phage-encoded SAM lyase revises catalytic function of enzyme family.
Elife, 10, 2021
|
|
8P2G
 
 | | Staphylococcus aureus 70S ribosome with elongation factor G locked with fusidic acid cyclopentane with a tRNA in pe/E chimeric state | | Descriptor: | 1,4-DIAMINOBUTANE, 16S ribosomal RNA, 2-[(3~{R},4~{S},5~{S},8~{S},9~{S},10~{S},11~{R},13~{S},14~{S},16~{S})-16-acetyloxy-4,8,10,14-tetramethyl-3,11-bis(oxidanyl)-1,2,3,4,5,6,7,9,11,12,13,15,16,17-tetradecahydrocyclopenta[a]phenanthren-17-yl]-5-cyclopentyl-pentanoic acid, ... | | Authors: | Gonzalez-Lopez, A, Selmer, M. | | Deposit date: | 2023-05-16 | | Release date: | 2024-05-01 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (2.02 Å) | | Cite: | Structures of the Staphylococcus aureus ribosome inhibited by fusidic acid and fusidic acid cyclopentane.
Sci Rep, 14, 2024
|
|
