4WOP
 
 | | Nucleotide Triphosphate Promiscuity in Mycobacterium tuberculosis Dethiobiotin Synthetase | | Descriptor: | ATP-dependent dethiobiotin synthetase BioD, CYTIDINE-5'-TRIPHOSPHATE, SULFATE ION | | Authors: | Salaemae, W, Yap, M.Y, Wegener, K.L, Booker, G.W, Wilce, M.C.J, Polyak, S.W. | | Deposit date: | 2014-10-16 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Nucleotide triphosphate promiscuity in Mycobacterium tuberculosis dethiobiotin synthetase.
Tuberculosis (Edinb), 95, 2015
|
|
6CZD
 
 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with adenosine diphosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent dethiobiotin synthetase BioD, MAGNESIUM ION | | Authors: | Thompson, A.P, Wegener, K.L, Bruning, J.B, Polyak, S.W. | | Deposit date: | 2018-04-09 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Precipitant-ligand exchange technique reveals the ADP binding mode in Mycobacterium tuberculosis dethiobiotin synthetase.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6CVF
 
 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with cytidine diphosphate | | Descriptor: | ATP-dependent dethiobiotin synthetase BioD, CYTIDINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Thompson, A.P, Bruning, J.B, Wegener, K.L, Polyak, S.W. | | Deposit date: | 2018-03-28 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mycobacterium tuberculosis Dethiobiotin Synthetase Facilitates Nucleoside Triphosphate Promiscuity through Alternate Binding Modes
Acs Catalysis, 8(11), 2018
|
|
6CZB
 
 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with uridine triphosphate (UTP) - promiscuous binding mode with disordered nucleoside | | Descriptor: | ATP-dependent dethiobiotin synthetase BioD, MAGNESIUM ION, URIDINE 5'-TRIPHOSPHATE | | Authors: | Thompson, A.P, Wegener, K.L, Bruning, J.B, Polyak, S.W. | | Deposit date: | 2018-04-09 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mycobacterium tuberculosis Dethiobiotin Synthetase Facilitates Nucleoside Triphosphate Promiscuity through Alternate Binding Modes
Acs Catalysis, 8(11), 2018
|
|
6CVE
 
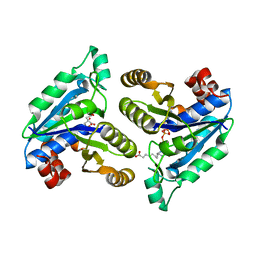 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin Synthetase in complex with cytidine triphosphate and 7,8-diaminopelargonic acid | | Descriptor: | 7-(CARBOXYAMINO)-8-AMINO-NONANOIC ACID, ATP-dependent dethiobiotin synthetase BioD, CITRATE ANION, ... | | Authors: | Thompson, A.P, Bruning, J.B, Wegener, K.L, Polyak, S.W. | | Deposit date: | 2018-03-28 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mycobacterium tuberculosis Dethiobiotin Synthetase Facilitates Nucleoside Triphosphate Promiscuity through Alternate Binding Modes
Acs Catalysis, 8(11), 2018
|
|
6CVV
 
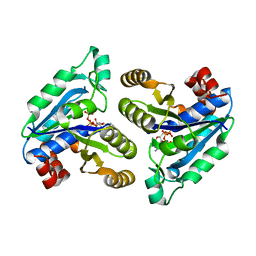 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with adenosine triphosphate (ATP) - promiscuous binding mode with disordered nucleoside | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent dethiobiotin synthetase BioD, MAGNESIUM ION | | Authors: | Thompson, A.P, Wegener, K.L, Bruning, J.B, Polyak, S.W. | | Deposit date: | 2018-03-29 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Mycobacterium tuberculosis Dethiobiotin Synthetase Facilitates Nucleoside Triphosphate Promiscuity through Alternate Binding Modes
Acs Catalysis, 8(11), 2018
|
|
6CZC
 
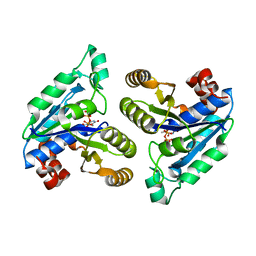 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with thymidine triphosphate (TTP) - promiscuous binding mode with disordered nucleoside | | Descriptor: | ATP-dependent dethiobiotin synthetase BioD, MAGNESIUM ION, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Thompson, A.P, Wegener, K.L, Bruning, J.B, Polyak, S.W. | | Deposit date: | 2018-04-09 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mycobacterium tuberculosis Dethiobiotin Synthetase Facilitates Nucleoside Triphosphate Promiscuity through Alternate Binding Modes
Acs Catalysis, 8(11), 2018
|
|
6CVU
 
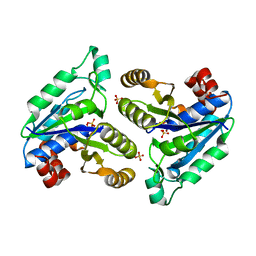 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with cytidine | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, ATP-dependent dethiobiotin synthetase BioD, SULFATE ION | | Authors: | Thompson, A.P, Wegener, K.L, Bruning, J.B, Polyak, S.W. | | Deposit date: | 2018-03-28 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.426 Å) | | Cite: | Mycobacterium tuberculosis Dethiobiotin Synthetase Facilitates Nucleoside Triphosphate Promiscuity through Alternate Binding Modes
Acs Catalysis, 8(11), 2018
|
|
6CZE
 
 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with inosine triphosphate (ITP) - promiscuous binding mode with disordered nucleoside | | Descriptor: | ATP-dependent dethiobiotin synthetase BioD, INOSINE 5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Thompson, A.P, Wegener, K.L, Bruning, J.B, Polyak, S.W. | | Deposit date: | 2018-04-09 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mycobacterium tuberculosis Dethiobiotin Synthetase Facilitates Nucleoside Triphosphate Promiscuity through Alternate Binding Modes
Acs Catalysis, 8(11), 2018
|
|
6E06
 
 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with cytidine triphosphate solved by precipitant-ligand exchange (crystals grown in citrate precipitant) | | Descriptor: | ATP-dependent dethiobiotin synthetase BioD, CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Thompson, A.P, Wegener, K.L, Bruning, J.B, Polyak, S.W. | | Deposit date: | 2018-07-06 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Precipitant-ligand exchange technique reveals the ADP binding mode in Mycobacterium tuberculosis dethiobiotin synthetase.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6E05
 
 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with cytidine triphosphate solved by precipitant-ligand exchange (crystals grown in sulfate precipitant) | | Descriptor: | ATP-dependent dethiobiotin synthetase BioD, CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Thompson, A.P, Wegener, K.L, Bruning, J.B, Polyak, S.W. | | Deposit date: | 2018-07-06 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Precipitant-ligand exchange technique reveals the ADP binding mode in Mycobacterium tuberculosis dethiobiotin synthetase.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6NDL
 
 | | Crystal structure of Staphylococcus aureus biotin protein ligase in complex with a sulfonamide inhibitor | | Descriptor: | 1-[4-(6-aminopurin-9-yl)butylsulfamoyl]-3-[4-[(4~{S})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]butyl]urea, Biotin Protein Ligase, GLYCEROL | | Authors: | Marshall, A.C, Polyak, S.W, Bruning, J.B, Lee, K. | | Deposit date: | 2018-12-13 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Sulfonamide-Based Inhibitors of Biotin Protein Ligase as New Antibiotic Leads.
Acs Chem.Biol., 14, 2019
|
|
6NMZ
 
 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with an intentionally produced fragment degradation product B9D | | Descriptor: | ATP-dependent dethiobiotin synthetase BioD, SULFATE ION, [(1R,2S)-2-(2-hydroxybenzene-1-carbonyl)cyclopentyl]acetic acid, ... | | Authors: | Thompson, A.P, Polyak, S.W, Wegener, K.L, Bruning, J.B. | | Deposit date: | 2019-01-14 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with an intentionally produced fragment degradation product B9D
To Be Published
|
|
6NVD
 
 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with triazole linked compound 9 | | Descriptor: | 2',5'-dideoxy-5'-[4-({[(1S,2R)-2-(2-hydroxybenzene-1-carbonyl)cyclopentyl]acetyl}amino)-1H-1,2,3-triazol-1-yl]cytidine, ATP-dependent dethiobiotin synthetase BioD, SULFATE ION | | Authors: | Thompson, A.P, Polyak, S.W, Wegener, K.L, Bruning, J.B. | | Deposit date: | 2019-02-05 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with triazole linked compound 9
To Be Published
|
|
6NKA
 
 | |
6NL5
 
 | |
6NVF
 
 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with fragment analogue 8 | | Descriptor: | (4-{[(1R,2R)-2-(carboxymethyl)cyclopentyl]methyl}phenyl)acetic acid, (4-{[(1S,2S)-2-(carboxymethyl)cyclopentyl]methyl}phenyl)acetic acid, ATP-dependent dethiobiotin synthetase BioD, ... | | Authors: | Thompson, A.P, Polyak, S.W, Wegener, K.L, Bruning, J.B. | | Deposit date: | 2019-02-05 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.989 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with fragment analogue 8
To Be Published
|
|
6NLZ
 
 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with fragment degradation product B9D | | Descriptor: | ATP-dependent dethiobiotin synthetase BioD, SULFATE ION, [(1R,2S)-2-(2-hydroxybenzene-1-carbonyl)cyclopentyl]acetic acid, ... | | Authors: | Thompson, A.P, Polyak, S.W, Wegener, K.L, Bruning, J.B. | | Deposit date: | 2019-01-10 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with fragment degradation product B9D
To Be Published
|
|
6NN0
 
 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with 2'-deoxycytidine and fragment degradation product B9D | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, ATP-dependent dethiobiotin synthetase BioD, SULFATE ION, ... | | Authors: | Thompson, A.P, Polyak, S.W, Wegener, K.L, Bruning, J.B. | | Deposit date: | 2019-01-14 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.343 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with 2'-deoxycytidine and B9D
To Be Published
|
|
6NKB
 
 | |
6NL4
 
 | |
6NWN
 
 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with glutamic acid linked compound 10 | | Descriptor: | 5'-{[N-({(1S,2R)-2-[4-(carboxymethyl)benzene-1-carbonyl]cyclopentyl}acetyl)-L-gamma-glutamyl]amino}-2',5'-dideoxycytidine, ATP-dependent dethiobiotin synthetase BioD, CITRIC ACID | | Authors: | Thompson, A.P, Polyak, S.W, Wegener, K.L, Bruning, J.B. | | Deposit date: | 2019-02-06 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with glutamic acid linked compound 10
To Be Published
|
|
6NWG
 
 | |
6NNZ
 
 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with fragment analogue 4 | | Descriptor: | ATP-dependent dethiobiotin synthetase BioD, SULFATE ION, [(1R,2S)-2-(benzenecarbonyl)cyclopentyl]acetic acid, ... | | Authors: | Thompson, A.P, Polyak, S.W, Wegener, K.L, Bruning, J.B. | | Deposit date: | 2019-01-15 | | Release date: | 2020-01-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with fragment analogue 4
To Be Published
|
|
6NVE
 
 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with fragment analogue 7 | | Descriptor: | 4-{[(1S,2S)-2-(carboxymethyl)cyclopentyl]methyl}benzoic acid, ATP-dependent dethiobiotin synthetase BioD, SULFATE ION | | Authors: | Thompson, A.P, Polyak, S.W, Wegener, K.L, Bruning, J.B. | | Deposit date: | 2019-02-05 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with fragment analogue 7
To Be Published
|
|
