1F88
 
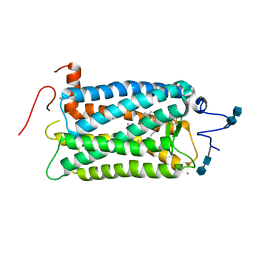 | | CRYSTAL STRUCTURE OF BOVINE RHODOPSIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MERCURY (II) ION, RETINAL, ... | | Authors: | Okada, T, Palczewski, K, Stenkamp, R.E, Miyano, M. | | Deposit date: | 2000-06-29 | | Release date: | 2000-08-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of rhodopsin: A G protein-coupled receptor.
Science, 289, 2000
|
|
8FD1
 
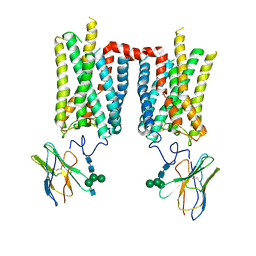 | | Crystal structure of photoactivated rhodopsin in complex with a nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody Nb2, ... | | Authors: | Salom, D, Palczewski, K, Kiser, P.D. | | Deposit date: | 2022-12-01 | | Release date: | 2023-08-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.25 Å) | | Cite: | Structural basis for the allosteric modulation of rhodopsin by nanobody binding to its extracellular domain.
Nat Commun, 14, 2023
|
|
8FCZ
 
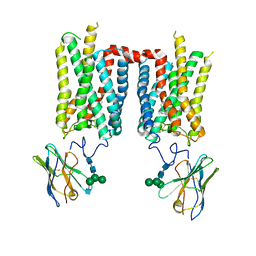 | | Crystal structure of ground-state rhodopsin in complex with a nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody Nb2, ... | | Authors: | Salom, D, Palczewski, K, Kiser, P.D. | | Deposit date: | 2022-12-01 | | Release date: | 2023-08-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural basis for the allosteric modulation of rhodopsin by nanobody binding to its extracellular domain.
Nat Commun, 14, 2023
|
|
8FD0
 
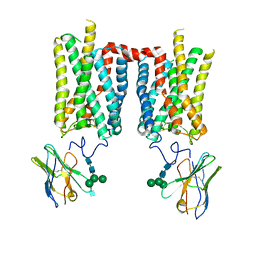 | | Crystal structure of bovine rod opsin in complex with a nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody Nb2, ... | | Authors: | Salom, D, Palczewski, K, Kiser, P.D. | | Deposit date: | 2022-12-01 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.71 Å) | | Cite: | Structural basis for the allosteric modulation of rhodopsin by nanobody binding to its extracellular domain.
Nat Commun, 14, 2023
|
|
6OFJ
 
 | |
4ZHK
 
 | | Crystal structure of RPE65 in complex with MB-002 | | Descriptor: | (1R)-1-[3-(cyclohexylmethoxy)phenyl]propane-1,3-diol, (1S)-1-[3-(cyclohexylmethoxy)phenyl]propane-1,3-diol, FE (II) ION, ... | | Authors: | Kiser, P.D, Palczewski, K. | | Deposit date: | 2015-04-25 | | Release date: | 2015-05-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Molecular pharmacodynamics of emixustat in protection against retinal degeneration.
J.Clin.Invest., 125, 2015
|
|
7MP2
 
 | | CRYSTAL STRUCTURE OF NATIVE BOVINE ARRESTIN 1 IN COMPLEX WITH 1D-MYO-INOSITOL 1,5-BISDIPHOSPHATE TETRAKISPHOSPHATE (1,5-PP IP4) | | Descriptor: | (1R,3S,4R,5S,6R)-2,4,5,6-tetrakis(phosphonooxy)cyclohexane-1,3-diyl bis[trihydrogen (diphosphate)], S-arrestin | | Authors: | Sander, C.L, Palczewski, K, Kiser, P.D. | | Deposit date: | 2021-05-04 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural evidence for visual arrestin priming via complexation of phosphoinositols.
Structure, 30, 2022
|
|
7MOR
 
 | |
7MP1
 
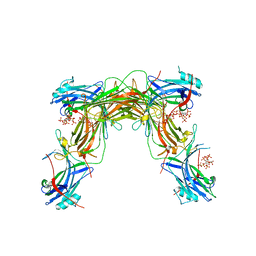 | | CRYSTAL STRUCTURE OF NATIVE BOVINE ARRESTIN 1 IN COMPLEX WITH 1,5-DI-METHYLENEBISPHOSPHONATE INOSITOL TETRAKISPHOSPHATE (1,5-PCP-IP4) | | Descriptor: | S-arrestin, {[(1R,3S,4S,5R,6S)-2,4,5,6-tetrakis(phosphonooxy)cyclohexane-1,3-diyl]bis[oxy(hydroxyphosphoryl)methanediyl]}bis(phosphonic acid) | | Authors: | Sander, C.L, Palczewski, K, Kiser, P.D. | | Deposit date: | 2021-05-04 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structural evidence for visual arrestin priming via complexation of phosphoinositols.
Structure, 30, 2022
|
|
7MP0
 
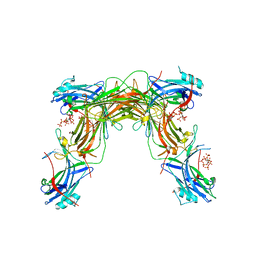 | | CRYSTAL STRUCTURE OF NATIVE BOVINE ARRESTIN 1 IN COMPLEX WITH 1D-MYO-INOSITOL 5-DIPHOSPHATE PENTAKISPHOSPHATE (5-PP IP5) | | Descriptor: | (1r,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, 2-ETHOXYETHANOL, S-arrestin | | Authors: | Sander, C.L, Palczewski, K, Kiser, P.D. | | Deposit date: | 2021-05-04 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural evidence for visual arrestin priming via complexation of phosphoinositols.
Structure, 30, 2022
|
|
6MZB
 
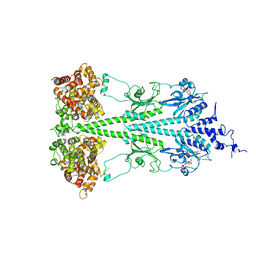 | | Cryo-EM structure of phosphodiesterase 6 | | Descriptor: | GUANOSINE-3',5'-MONOPHOSPHATE, MAGNESIUM ION, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit gamma, ... | | Authors: | Gulati, S, Palczewski, K. | | Deposit date: | 2018-11-04 | | Release date: | 2019-03-06 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of phosphodiesterase 6 reveals insights into the allosteric regulation of type I phosphodiesterases.
Sci Adv, 5, 2019
|
|
1HZX
 
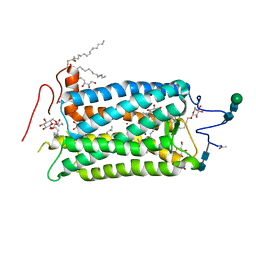 | | CRYSTAL STRUCTURE OF BOVINE RHODOPSIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HEPTANE-1,2,3-TRIOL, MERCURY (II) ION, ... | | Authors: | Teller, D.C, Okada, T, Behnke, C.A, Palczewski, K, Stenkamp, R.E. | | Deposit date: | 2001-01-26 | | Release date: | 2001-07-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Advances in determination of a high-resolution three-dimensional structure of rhodopsin, a model of G-protein-coupled receptors (GPCRs).
Biochemistry, 40, 2001
|
|
2NWC
 
 | |
1JBA
 
 | | UNMYRISTOYLATED GCAP-2 WITH THREE CALCIUM IONS BOUND | | Descriptor: | CALCIUM ION, PROTEIN (GUANYLATE CYCLASE ACTIVATING PROTEIN 2) | | Authors: | Ames, J.B, Dizhoor, A.M, Ikura, M, Palczewski, K, Stryer, L. | | Deposit date: | 1999-04-03 | | Release date: | 1999-12-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of guanylyl cyclase activating protein-2, a calcium-sensitive modulator of photoreceptor guanylyl cyclases.
J.Biol.Chem., 274, 1999
|
|
3FSN
 
 | | Crystal structure of RPE65 at 2.14 angstrom resolution | | Descriptor: | FE (II) ION, Retinal pigment epithelium-specific 65 kDa protein, TETRAETHYLENE GLYCOL | | Authors: | Kiser, P.D, Lodowski, D.T, Palczewski, K. | | Deposit date: | 2009-01-11 | | Release date: | 2009-09-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of native RPE65, the retinoid isomerase of the visual cycle.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3KVC
 
 | | Crystal structure of bovine RPE65 at 1.9 angstrom resolution | | Descriptor: | FE (II) ION, Retinoid isomerohydrolase | | Authors: | Kiser, P.D, Lodowski, D.T, Golczak, M, Palczewski, K. | | Deposit date: | 2009-11-29 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Importance of membrane structural integrity for RPE65 retinoid isomerization activity.
J.Biol.Chem., 285, 2010
|
|
3LM1
 
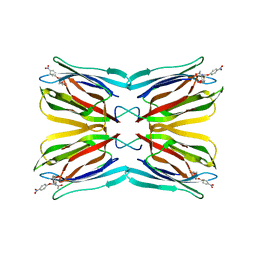 | | Crystal Structure Analysis of Maclura pomifera agglutinin complex with p-nitrophenyl-GalNAc | | Descriptor: | 4-nitrophenyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, Agglutinin alpha chain, Agglutinin beta-2 chain | | Authors: | Huang, J, Xu, Z, Wang, D, Ogato, C, Hirama, T, Palczewski, K, Hazen, S.L, Lee, X, Young, N.M. | | Deposit date: | 2010-01-29 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of the secondary binding sites of Maclura pomifera agglutinin by glycan array and crystallographic analyses.
Glycobiology, 20, 2010
|
|
5KJD
 
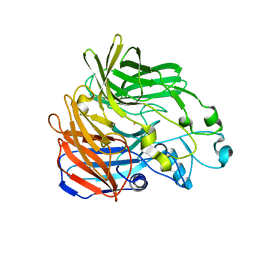 | |
5KK0
 
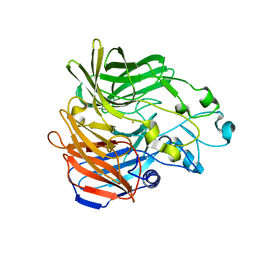 | | Synechocystis ACO mutant - T136A | | Descriptor: | Apocarotenoid-15,15'-oxygenase, CHLORIDE ION, FE (II) ION | | Authors: | Sui, X, Kiser, P.D, Palczewski, K. | | Deposit date: | 2016-06-20 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Utilization of Dioxygen by Carotenoid Cleavage Oxygenases.
J.Biol.Chem., 290, 2015
|
|
5KJB
 
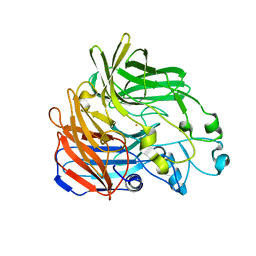 | |
5KJA
 
 | | Synechocystis apocarotenoid oxygenase (ACO) mutant - Trp149Ala | | Descriptor: | Apocarotenoid-15,15'-oxygenase, CHLORIDE ION, FE (II) ION | | Authors: | Sui, X, Kiser, P.D, Palczewski, K. | | Deposit date: | 2016-06-18 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Key Residues for Catalytic Function and Metal Coordination in a Carotenoid Cleavage Dioxygenase.
J.Biol.Chem., 291, 2016
|
|
3VAT
 
 | | Crystal structure of DNPEP, ZnMg form | | Descriptor: | Aspartyl aminopeptidase, MAGNESIUM ION, ZINC ION | | Authors: | Kiser, P.D, Chen, Y, Palczewski, K. | | Deposit date: | 2011-12-29 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights into substrate specificity and metal activation of Mammalian tetrahedral aspartyl aminopeptidase.
J.Biol.Chem., 287, 2012
|
|
3VAR
 
 | | Crystal structure of DNPEP, ZnZn form | | Descriptor: | Aspartyl aminopeptidase, ZINC ION | | Authors: | Kiser, P.D, Chen, Y, Palczewski, K. | | Deposit date: | 2011-12-29 | | Release date: | 2012-02-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Insights into substrate specificity and metal activation of Mammalian tetrahedral aspartyl aminopeptidase.
J.Biol.Chem., 287, 2012
|
|
5TE5
 
 | | Crystal structure of Bos taurus opsin regenerated with 6-carbon ring retinal chromophore | | Descriptor: | (2E)-{(4E)-4-[(3E)-4-(2,6,6-trimethylcyclohex-1-en-1-yl)but-3-en-2-ylidene]cyclohex-2-en-1-ylidene}acetaldehyde, Rhodopsin | | Authors: | Gulati, S, Banerjee, S, Katayama, K, Kiser, P.D, Palczewski, K. | | Deposit date: | 2016-09-20 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.01 Å) | | Cite: | Photocyclic behavior of rhodopsin induced by an atypical isomerization mechanism.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3LLY
 
 | | Crystal Structure Analysis of Maclura pomifera agglutinin | | Descriptor: | Agglutinin alpha chain, Agglutinin beta-2 chain | | Authors: | Huang, J, Xu, Z, Wang, D, Ogato, C, Hirama, T, Palczewski, K, Hazen, S.L, Lee, X, Young, N.M. | | Deposit date: | 2010-01-29 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Characterization of the secondary binding sites of Maclura pomifera agglutinin by glycan array and crystallographic analyses.
Glycobiology, 20, 2010
|
|
