6RQT
 
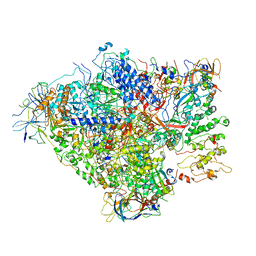 | | RNA Polymerase I-tWH-Rrn3-DNA | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Mueller, C.W, Sadian, Y, Tafur, L. | | Deposit date: | 2019-05-16 | | Release date: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | RNA Polymerase I-tWH-Rrn3-DNA
To Be Published
|
|
1AKE
 
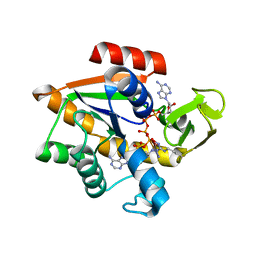 | |
1E4Y
 
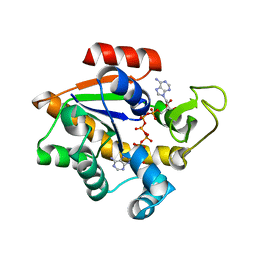 | |
1E4V
 
 | |
4C5H
 
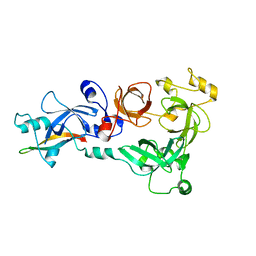 | |
4C5E
 
 | |
4C5I
 
 | | Crystal structure of MBTD1 YY1 complex | | Descriptor: | MBT DOMAIN-CONTAINING PROTEIN 1, TRANSCRIPTIONAL REPRESSOR PROTEIN YY1 | | Authors: | Alfieri, C, Glatt, S, Mueller, C.W. | | Deposit date: | 2013-09-12 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.586 Å) | | Cite: | Structural Basis for Targeting the Chromatin Repressor Sfmbt to Polycomb Response Elements
Genes Dev., 27, 2013
|
|
4C5G
 
 | |
2WP2
 
 | | Structure of Brdt bromodomain BD1 bound to a diacetylated histone H4 peptide. | | Descriptor: | BROMODOMAIN TESTIS-SPECIFIC PROTEIN, HISTONE H4 | | Authors: | Moriniere, J, Rousseaux, S, Steuerwald, U, Soler-Lopez, M, Curtet, S, Vitte, A.-L, Govin, J, Gaucher, J, Sadoul, K, Hart, D.J, Krijgsveld, J, Khochbin, S, Mueller, C.W, Petosa, C. | | Deposit date: | 2009-08-02 | | Release date: | 2009-09-22 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Cooperative Binding of Two Acetylation Marks on a Histone Tail by a Single Bromodomain.
Nature, 461, 2009
|
|
2WP1
 
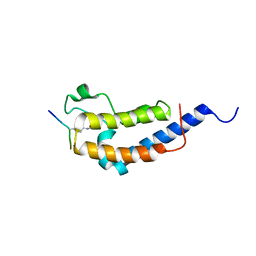 | | Structure of Brdt bromodomain 2 bound to an acetylated histone H3 peptide | | Descriptor: | BROMODOMAIN TESTIS-SPECIFIC PROTEIN, HISTONE H3 | | Authors: | Moriniere, J, Rousseaux, S, Steuerwald, U, Soler-Lopez, M, Curtet, S, Vitte, A.-L, Govin, J, Gaucher, J, Sadoul, K, Hart, D.J, Krijgsveld, J, Khochbin, S, Mueller, C.W, Petosa, C. | | Deposit date: | 2009-08-02 | | Release date: | 2009-09-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cooperative Binding of Two Acetylation Marks on a Histone Tail by a Single Bromodomain.
Nature, 461, 2009
|
|
1SVC
 
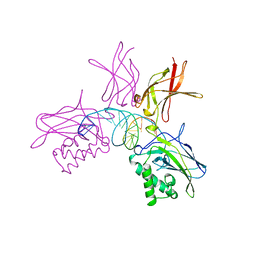 | | NFKB P50 HOMODIMER BOUND TO DNA | | Descriptor: | DNA (5'-D(*AP*GP*AP*TP*GP*GP*GP*GP*AP*AP*TP*CP*CP*CP*CP*TP*A P*GP*A)-3'), PROTEIN (NUCLEAR FACTOR KAPPA-B (NF-KB)) | | Authors: | Mueller, C.W, Harrison, S.C. | | Deposit date: | 1995-11-27 | | Release date: | 1996-06-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the NF-kappa B p50 homodimer bound to DNA.
Nature, 373, 1995
|
|
6RRD
 
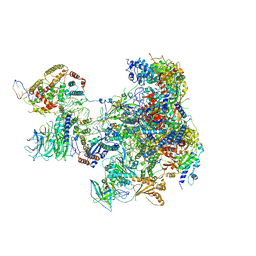 | | RNA Polymerase I Pre-initiation complex DNA opening intermediate 1 | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Mueller, C.W, Sadian, Y, Tafur, L. | | Deposit date: | 2019-05-17 | | Release date: | 2019-12-11 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular insight into RNA polymerase I promoter recognition and promoter melting.
Nat Commun, 10, 2019
|
|
6RUO
 
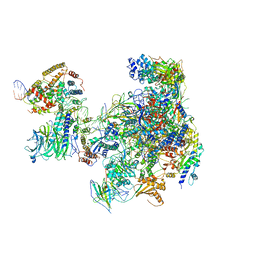 | | RNA Polymerase I Open Complex conformation 1 | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Mueller, C.W, Sadian, Y, Tafur, L. | | Deposit date: | 2019-05-28 | | Release date: | 2019-12-11 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular insight into RNA polymerase I promoter recognition and promoter melting.
Nat Commun, 10, 2019
|
|
6RUI
 
 | | RNA Polymerase I Pre-initiation complex DNA opening intermediate 2 | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Mueller, C.W, Sadian, Y, Tafur, L. | | Deposit date: | 2019-05-28 | | Release date: | 2019-12-11 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Molecular insight into RNA polymerase I promoter recognition and promoter melting.
Nat Commun, 10, 2019
|
|
6RQL
 
 | | RNA Polymerase I Closed Conformation 2 (CC2) | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Mueller, C.W, Sadian, Y, Tafur, L. | | Deposit date: | 2019-05-16 | | Release date: | 2019-12-11 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular insight into RNA polymerase I promoter recognition and promoter melting.
Nat Commun, 10, 2019
|
|
6RQH
 
 | | RNA Polymerase I Closed Conformation 1 (CC1) | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Mueller, C.W, Sadian, Y, Tafur, L. | | Deposit date: | 2019-05-15 | | Release date: | 2019-12-11 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular insight into RNA polymerase I promoter recognition and promoter melting.
Nat Commun, 10, 2019
|
|
6RWE
 
 | | RNA Polymerase I Open Complex conformation 2 | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Mueller, C.W, Sadian, Y, Tafur, L. | | Deposit date: | 2019-06-04 | | Release date: | 2019-12-11 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular insight into RNA polymerase I promoter recognition and promoter melting.
Nat Commun, 10, 2019
|
|
1BVO
 
 | | DORSAL HOMOLOGUE GAMBIF1 BOUND TO DNA | | Descriptor: | DNA DUPLEX, TRANSCRIPTION FACTOR GAMBIF1 | | Authors: | Cramer, P, Varrot, A, Barillas-Mury, C, Kafatos, F.C, Mueller, C.W. | | Deposit date: | 1998-09-16 | | Release date: | 1999-07-12 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the specificity domain of the Dorsal homologue Gambif1 bound to DNA.
Structure Fold.Des., 7, 1999
|
|
8CLI
 
 | | TFIIIC TauB-DNA monomer | | Descriptor: | DNA (35-MER), General transcription factor 3C polypeptide 1, General transcription factor 3C polypeptide 2, ... | | Authors: | Seifert-Davila, W, Girbig, M, Hauptmann, L, Hoffmann, T, Eustermann, S, Mueller, C.W. | | Deposit date: | 2023-02-16 | | Release date: | 2023-06-21 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into human TFIIIC promoter recognition.
Sci Adv, 9, 2023
|
|
8CLK
 
 | | TFIIIC TauA complex | | Descriptor: | General transcription factor 3C polypeptide 1, General transcription factor 3C polypeptide 3, General transcription factor 3C polypeptide 5, ... | | Authors: | Seifert-Davila, W, Girbig, M, Hauptmann, L, Hoffmann, T, Eustermann, S, Mueller, C.W. | | Deposit date: | 2023-02-16 | | Release date: | 2023-06-21 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into human TFIIIC promoter recognition.
Sci Adv, 9, 2023
|
|
8CLL
 
 | | Structural insights into human TFIIIC promoter recognition | | Descriptor: | General transcription factor 3C polypeptide 1, General transcription factor 3C polypeptide 2, General transcription factor 3C polypeptide 4, ... | | Authors: | Seifert-Davila, W, Girbig, M, Hauptmann, L, Hoffmann, T, Eustermann, S, Mueller, C.W. | | Deposit date: | 2023-02-16 | | Release date: | 2023-06-21 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into human TFIIIC promoter recognition.
Sci Adv, 9, 2023
|
|
8CLJ
 
 | | TFIIIC TauB-DNA dimer | | Descriptor: | General transcription factor 3C polypeptide 1, General transcription factor 3C polypeptide 2, General transcription factor 3C polypeptide 4, ... | | Authors: | Seifert-Davila, W, Girbig, M, Hauptmann, L, Hoffmann, T, Eustermann, S, Mueller, C.W. | | Deposit date: | 2023-02-16 | | Release date: | 2023-06-21 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into human TFIIIC promoter recognition.
Sci Adv, 9, 2023
|
|
6TUT
 
 | | Cryo-EM structure of the RNA Polymerase III-Maf1 complex | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Vorlaender, M.K, Hagen, W.J.H, Mueller, C.W. | | Deposit date: | 2020-01-08 | | Release date: | 2020-02-19 | | Last modified: | 2020-03-18 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural basis for RNA polymerase III transcription repression by Maf1.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7Z1O
 
 | | Structure of yeast RNA Polymerase III PTC + NTPs | | Descriptor: | CHAPSO, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Girbig, M, Mueller, C.W. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-31 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Architecture of the yeast Pol III pre-termination complex and pausing mechanism on poly(dT) termination signals.
Cell Rep, 40, 2022
|
|
7Z1N
 
 | | Structure of yeast RNA Polymerase III Delta C53-C37-C11 | | Descriptor: | CHAPSO, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Girbig, M, Mueller, C.W. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-31 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Architecture of the yeast Pol III pre-termination complex and pausing mechanism on poly(dT) termination signals.
Cell Rep, 40, 2022
|
|
