1IS7
 
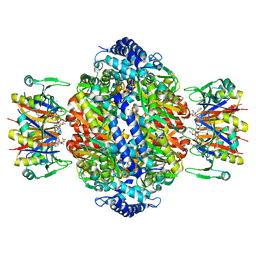 | | Crystal structure of rat GTPCHI/GFRP stimulatory complex | | Descriptor: | GTP Cyclohydrolase I, GTP Cyclohydrolase I Feedback Regulatory Protein, PHENYLALANINE, ... | | Authors: | Maita, N, Okada, K, Hatakeyama, K, Hakoshima, T. | | Deposit date: | 2001-11-18 | | Release date: | 2002-02-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the stimulatory complex of GTP cyclohydrolase I and its feedback regulatory protein GFRP.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1IS8
 
 | | Crystal structure of rat GTPCHI/GFRP stimulatory complex plus Zn | | Descriptor: | GTP Cyclohydrolase I, GTP Cyclohydrolase I Feedback Regulatory Protein, PHENYLALANINE, ... | | Authors: | Maita, N, Okada, K, Hatakeyama, K, Hakoshima, T. | | Deposit date: | 2001-11-18 | | Release date: | 2002-02-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the stimulatory complex of GTP cyclohydrolase I and its feedback regulatory protein GFRP.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2EI9
 
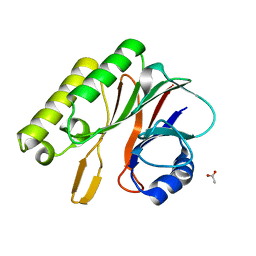 | | Crystal structure of R1Bm endonuclease domain | | Descriptor: | ACETIC ACID, Non-LTR retrotransposon R1Bmks ORF2 protein | | Authors: | Maita, N, Aoyagi, H, Osanai, M, Shirakawa, M, Fujiwara, H. | | Deposit date: | 2007-03-12 | | Release date: | 2007-07-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of the sequence specificity of the R1Bm endonuclease domain by structural and biochemical studies.
Nucleic Acids Res., 35, 2007
|
|
6A43
 
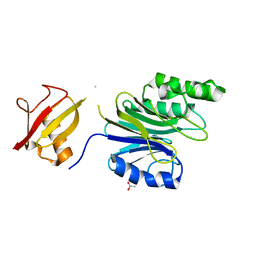 | | R1EN(5-225)-ubiquitin fusion | | Descriptor: | ACETIC ACID, RNA-directed DNA polymerase homolog (R1),Polyubiquitin-C | | Authors: | Maita, N. | | Deposit date: | 2018-06-19 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure Determination of Ubiquitin by Fusion to a Protein That Forms a Highly Porous Crystal Lattice
J. Am. Chem. Soc., 140, 2018
|
|
6A44
 
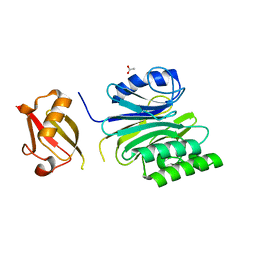 | | R1EN(5-227)-ubiquitin fusion | | Descriptor: | ACETIC ACID, RNA-directed DNA polymerase homolog (R1),Polyubiquitin-C | | Authors: | Maita, N. | | Deposit date: | 2018-06-19 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure Determination of Ubiquitin by Fusion to a Protein That Forms a Highly Porous Crystal Lattice
J. Am. Chem. Soc., 140, 2018
|
|
6A42
 
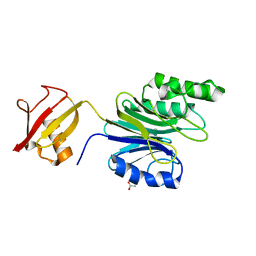 | | R1EN(5-223)-ubiquitin fusion | | Descriptor: | ACETIC ACID, RNA-directed DNA polymerase homolog (R1),Polyubiquitin-C | | Authors: | Maita, N. | | Deposit date: | 2018-06-19 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure Determination of Ubiquitin by Fusion to a Protein That Forms a Highly Porous Crystal Lattice
J. Am. Chem. Soc., 140, 2018
|
|
1UEX
 
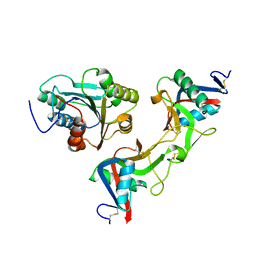 | | Crystal structure of von Willebrand Factor A1 domain complexed with snake venom bitiscetin | | Descriptor: | bitiscetin alpha chain, bitiscetin beta chain, von Willebrand Factor | | Authors: | Maita, N, Nishio, K, Nishimoto, E, Matsui, T, Shikamoto, Y, Morita, T, Sadler, J.E, Mizuno, H. | | Deposit date: | 2003-05-22 | | Release date: | 2003-09-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of von Willebrand factor A1 domain complexed with snake venom, bitiscetin. Insight into glycoprotein Ibalpha binding mechanism induced by snake venom proteins.
J.Biol.Chem., 278, 2003
|
|
1WDU
 
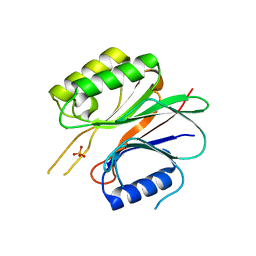 | | Endonuclease domain of TRAS1, a telomere-specific non-LTR retrotransposon | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, TRAS1 ORF2p | | Authors: | Maita, N, Anzai, T, Aoyagi, H, Mizuno, H, Fujiwara, H. | | Deposit date: | 2004-05-17 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the endonuclease domain encoded by the telomere-specific long interspersed nuclear element, TRAS1
J.Biol.Chem., 279, 2004
|
|
1WPL
 
 | | Crystal structure of the inhibitory form of rat GTP cyclohydrolase I/GFRP complex | | Descriptor: | 7,8-DIHYDROBIOPTERIN, GTP cyclohydrolase I, GTP cyclohydrolase I feedback regulatory protein, ... | | Authors: | Maita, N, Hatakeyama, K, Okada, K, Hakoshima, T. | | Deposit date: | 2004-09-08 | | Release date: | 2004-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of biopterin-induced inhibition of GTP cyclohydrolase I by GFRP, its feedback regulatory protein
J.Biol.Chem., 279, 2004
|
|
3W81
 
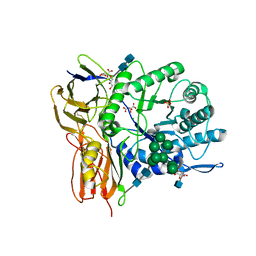 | | Human alpha-l-iduronidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-L-iduronidase, ... | | Authors: | Maita, N, Tsukimura, T, Taniguchi, T, Saito, S, Ohno, K, Taniguchi, H, Sakuraba, H. | | Deposit date: | 2013-03-11 | | Release date: | 2013-08-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Human alpha-L-iduronidase uses its own N-glycan as a substrate-binding and catalytic module
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3W82
 
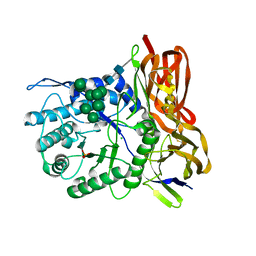 | | Human alpha-L-iduronidase in complex with iduronic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-L-iduronidase, ... | | Authors: | Maita, N, Tsukimura, T, Taniguchi, T, Saito, S, Ohno, K, Taniguchi, H, Sakuraba, H. | | Deposit date: | 2013-03-11 | | Release date: | 2013-08-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Human alpha-L-iduronidase uses its own N-glycan as a substrate-binding and catalytic module
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3AAG
 
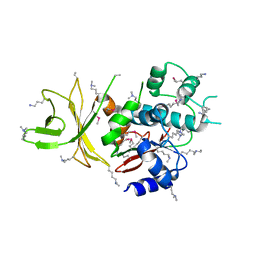 | | Crystal structure of C. jejuni pglb C-terminal domain | | Descriptor: | CALCIUM ION, General glycosylation pathway protein | | Authors: | Maita, N, Kohda, D. | | Deposit date: | 2009-11-16 | | Release date: | 2009-12-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Comparative structural biology of Eubacterial and Archaeal oligosaccharyltransferases.
J.Biol.Chem., 285, 2010
|
|
7X4O
 
 | |
4Z2L
 
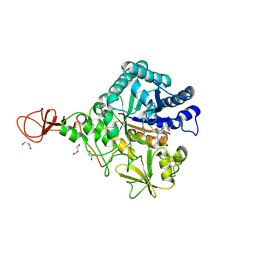 | | Serratia marcescens Chitinase B complexed with macrolide inhibitor 33 | | Descriptor: | (3R,4S,5S,6R,7R,9R,10S,11S,12R,13S,14R)-14-ethyl-7,10,12,13-tetrahydroxy-3,5,7,9,11,13-hexamethyl-2-oxo-6-(prop-2-yn-1-yloxy)oxacyclotetradecan-4-yl {3-[N'-(methylcarbamoyl)carbamimidamido]propyl}carbamate, CHLORIDE ION, Chitinase B, ... | | Authors: | Maita, N, Sugawara, A, Sunazuka, T. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Creation of Customized Bioactivity within a 14-Membered Macrolide Scaffold: Design, Synthesis, and Biological Evaluation Using a Family-18 Chitinase
J.Med.Chem., 58, 2015
|
|
4Z2G
 
 | | Serratia marcescens Chitinase B complexed with macrolide inhibitor 26 | | Descriptor: | (1R,2R,3R,6R,7S,8S,9R,10R,12R,13S,17S)-3-ethyl-2,10-dihydroxy-2,6,8,10,12,15,15,17-octamethyl-5-oxo-9-(prop-2-yn-1-yloxy)-4,14,16-trioxabicyclo[11.3.1]heptadec-7-yl {4-[N'-(methylcarbamoyl)carbamimidamido]butyl}carbamate, Chitinase B | | Authors: | Maita, N, Sugawara, A, Sunazuka, T. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Creation of Customized Bioactivity within a 14-Membered Macrolide Scaffold: Design, Synthesis, and Biological Evaluation Using a Family-18 Chitinase
J.Med.Chem., 58, 2015
|
|
4Z2J
 
 | | Serratia marcescens Chitinase B complexed with macrolide inhibitor 31 | | Descriptor: | (1R,2R,3R,6R,7S,8S,9R,10R,12R,13S,17S)-3-ethyl-2,10-dihydroxy-2,6,8,10,12,15,15,17-octamethyl-5-oxo-9-(prop-2-yn-1-yloxy)-4,14,16-trioxabicyclo[11.3.1]heptadec-7-yl {5-[N'-(methylcarbamoyl)carbamimidamido]pentyl}carbamate, Chitinase B, GLYCEROL, ... | | Authors: | Maita, N, Sugawara, A, Sunazuka, T. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Creation of Customized Bioactivity within a 14-Membered Macrolide Scaffold: Design, Synthesis, and Biological Evaluation Using a Family-18 Chitinase
J.Med.Chem., 58, 2015
|
|
4Z2K
 
 | | Serratia marcescens Chitinase B complexed with macrolide inhibitor 32 | | Descriptor: | (1R,2R,3R,6R,7S,8S,9R,10R,12R,13S,17S)-3-ethyl-2,10-dihydroxy-2,6,8,10,12,15,15,17-octamethyl-5-oxo-9-(prop-2-yn-1-yloxy)-4,14,16-trioxabicyclo[11.3.1]heptadec-7-yl {7-[N'-(methylcarbamoyl)carbamimidamido]heptyl}carbamate, Chitinase B, GLYCEROL | | Authors: | Maita, N, Sugawara, A, Sunazuka, T. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Creation of Customized Bioactivity within a 14-Membered Macrolide Scaffold: Design, Synthesis, and Biological Evaluation Using a Family-18 Chitinase
J.Med.Chem., 58, 2015
|
|
4Z2H
 
 | | Serratia marcescens Chitinase B complexed with macrolide inhibitor 29 | | Descriptor: | (1R,2R,3R,6R,7S,8S,9R,10R,12R,13S,17S)-3-ethyl-2,10-dihydroxy-2,6,8,10,12,15,15,17-octamethyl-5-oxo-9-(prop-2-yn-1-yloxy)-4,14,16-trioxabicyclo[11.3.1]heptadec-7-yl {2-[N'-(methylcarbamoyl)carbamimidamido]ethyl}carbamate, CHLORIDE ION, Chitinase B, ... | | Authors: | Maita, N, Sugawara, A, Sunazuka, T. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Creation of Customized Bioactivity within a 14-Membered Macrolide Scaffold: Design, Synthesis, and Biological Evaluation Using a Family-18 Chitinase
J.Med.Chem., 58, 2015
|
|
4Z2I
 
 | | Serratia marcescens Chitinase B complexed with macrolide inhibitor 30 | | Descriptor: | (1R,2R,3R,6R,7S,8S,9R,10R,12R,13S,17S)-3-ethyl-2,10-dihydroxy-2,6,8,10,12,15,15,17-octamethyl-5-oxo-9-(prop-2-yn-1-yloxy)-4,14,16-trioxabicyclo[11.3.1]heptadec-7-yl {3-[N'-(methylcarbamoyl)carbamimidamido]propyl}carbamate, Chitinase B, GLYCEROL, ... | | Authors: | Maita, N, Sugawara, A, Sunazuka, T. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Creation of Customized Bioactivity within a 14-Membered Macrolide Scaffold: Design, Synthesis, and Biological Evaluation Using a Family-18 Chitinase
J.Med.Chem., 58, 2015
|
|
2ZAG
 
 | |
2ZAI
 
 | | Crystal structure of the soluble domain of STT3 from P. furiosus | | Descriptor: | CALCIUM ION, CHLORIDE ION, Oligosaccharyl transferase stt3 subunit related protein | | Authors: | Maita, N. | | Deposit date: | 2007-10-05 | | Release date: | 2007-12-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-guided identification of a new catalytic motif of oligosaccharyltransferase
Embo J., 27, 2008
|
|
8YBF
 
 | | Crystal structure of canine distemper virus hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin glycoprotein | | Authors: | Fukuhara, H, Yumoto, K, Sako, M, Kajikawa, M, Ose, T, Hashiguchi, T, Kamishikiryo, J, Maita, N, Kuroki, K, Maenaka, K. | | Deposit date: | 2024-02-13 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Glycan-shielded homodimer structure and dynamical features of the canine distemper virus hemagglutinin relevant for viral entry and efficient vaccination.
Elife, 12, 2024
|
|
3VU0
 
 | | Crystal structure of the C-terminal globular domain of oligosaccharyltransferase (AfAglB-S2, AF_0040, O30195_ARCFU) from Archaeoglobus fulgidus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Putative uncharacterized protein | | Authors: | Nyirenda, J, Matsumoto, S, Saitoh, T, Maita, N, Noda, N.N, Inagaki, F, Kohda, D. | | Deposit date: | 2012-06-13 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystallographic and NMR Evidence for Flexibility in Oligosaccharyltransferases and Its Catalytic Significance
Structure, 21, 2013
|
|
7DMM
 
 | |
1ISN
 
 | | Crystal structure of merlin FERM domain | | Descriptor: | merlin | | Authors: | Shimizu, T, Seto, A, Maita, N, Hamada, K, Tsukita, S, Tsukita, S, Hakoshima, T. | | Deposit date: | 2001-12-13 | | Release date: | 2002-04-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for neurofibromatosis type 2. Crystal structure of the merlin FERM domain.
J.Biol.Chem., 277, 2002
|
|
