5JJT
 
 | |
1SNB
 
 | | STRUCTURE OF SCORPION NEUROTOXIN BMK M8 | | Descriptor: | NEUROTOXIN BMK M8 | | Authors: | Wang, D.C, Zeng, Z.H, Li, H.M. | | Deposit date: | 1997-03-12 | | Release date: | 1997-05-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an acidic neurotoxin from scorpion Buthus martensii Karsch at 1.85 A resolution.
J.Mol.Biol., 261, 1996
|
|
1B9E
 
 | | HUMAN INSULIN MUTANT SERB9GLU | | Descriptor: | PROTEIN (INSULIN) | | Authors: | Wang, D.C, Zeng, Z.H, Yao, Z.P, Li, H.M. | | Deposit date: | 1998-11-12 | | Release date: | 1999-11-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of an insulin dimer in an orthorhombic crystal: the structure analysis of a human insulin mutant (B9 Ser-->Glu).
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1GIK
 
 | | POKEWEED ANTIVIRAL PROTEIN FROM SEEDS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIVIRAL PROTEIN S | | Authors: | Zeng, Z.H, He, X.L, Li, H.M, Hu, Z, Wang, D.C. | | Deposit date: | 2001-02-07 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of pokeweed antiviral protein with well-defined sugars from seeds at 1.8 angstrom resolution
J.Struct.Biol., 141, 2003
|
|
5I0A
 
 | | RecA mini intein in complex with cisplatin | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, Cisplatin, Intein, ... | | Authors: | Li, Z, Zhang, J, Li, H.M. | | Deposit date: | 2016-02-03 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into protein splicing inhibition by platinum therapeutics as potential anti-microbials
To be published
|
|
5K08
 
 | | RecA mini intein-Zeise's salt complex | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, RecA mini intein, SULFATE ION, ... | | Authors: | Li, Z, Li, H.M. | | Deposit date: | 2016-05-17 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Structural insights into protein splicing inhibition by platinum therapeutics as potential anti-microbials
To be published
|
|
1SN4
 
 | | STRUCTURE OF SCORPION NEUROTOXIN BMK M4 | | Descriptor: | ACETATE ION, PROTEIN (NEUROTOXIN BMK M4) | | Authors: | He, X.L, Li, H.M, Liu, X.Q, Zeng, Z.H, Wang, D.C. | | Deposit date: | 1998-11-11 | | Release date: | 1999-11-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structures of two alpha-like scorpion toxins: non-proline cis peptide bonds and implications for new binding site selectivity on the sodium channel.
J.Mol.Biol., 292, 1999
|
|
1SN1
 
 | | STRUCTURE OF SCORPION NEUROTOXIN BMK M1 | | Descriptor: | PROTEIN (NEUROTOXIN BMK M1) | | Authors: | He, X.L, Li, H.M, Liu, X.Q, Zeng, Z.H, Wang, D.C. | | Deposit date: | 1998-11-12 | | Release date: | 1999-11-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of two alpha-like scorpion toxins: non-proline cis peptide bonds and implications for new binding site selectivity on the sodium channel.
J.Mol.Biol., 292, 1999
|
|
1H65
 
 | | Crystal structure of pea Toc34 - a novel GTPase of the chloroplast protein translocon | | Descriptor: | CHLOROPLAST OUTER ENVELOPE PROTEIN OEP34, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Sun, Y.J, Forouhar, F, Li, H.M, Tu, S.L, Kao, S, Shr, H.L, Chou, C.C, Hsiao, C.D. | | Deposit date: | 2001-06-06 | | Release date: | 2002-01-29 | | Last modified: | 2019-06-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Pea Toc34 - a Novel Gtpase of the Chloroplast Protein Translocon
Nat.Struct.Biol., 9, 2002
|
|
1CHZ
 
 | | A NEW NEUROTOXIN FROM BUTHUS MARTENSII KARSCH | | Descriptor: | CHLORIDE ION, PROTEIN (BMK M2) | | Authors: | He, X.L, Deng, J.P, Li, H.M, Wang, D.C. | | Deposit date: | 1999-03-31 | | Release date: | 2000-03-31 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure of a new neurotoxin from the scorpion Buthus martensii Karsch at 1.76 A.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
2OY0
 
 | | Crystal structure of the West Nile virus methyltransferase | | Descriptor: | Methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Li, H.M, Zhao, Y.W, Guo, Y, Shi, P.Y. | | Deposit date: | 2007-02-21 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and Function of Flavivirus NS5 Methyltransferase.
J.Virol., 81, 2007
|
|
1TVD
 
 | | VARIABLE DOMAIN OF T CELL RECEPTOR DELTA CHAIN | | Descriptor: | T CELL RECEPTOR | | Authors: | Li, H.M, Lebedeva, M.I, Llera, A.S, Fields, B.A, Brenner, M.B, Mariuzza, R.A. | | Deposit date: | 1997-10-19 | | Release date: | 1998-04-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Vdelta domain of a human gammadelta T-cell antigen receptor.
Nature, 391, 1998
|
|
4R05
 
 | | Crystal structure of the refolded DENV3 methyltransferase | | Descriptor: | Nonstructural protein NS5 | | Authors: | Brecher, M.B, Li, Z, Zhang, J, Chen, H, Lin, Q, Liu, B, Li, H.M. | | Deposit date: | 2014-07-29 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Refolding of a fully functional flavivirus methyltransferase revealed that S-adenosyl methionine but not S-adenosyl homocysteine is copurified with flavivirus methyltransferase.
Protein Sci., 24, 2015
|
|
3KPH
 
 | |
3LKZ
 
 | |
4IQ2
 
 | | P21 crystal form of FKBP12.6 | | Descriptor: | MALONIC ACID, Peptidyl-prolyl cis-trans isomerase FKBP1B | | Authors: | Chen, H, Mustafi, S.M, Li, H.M, LeMaster, D.M, Hernandez, G. | | Deposit date: | 2013-01-10 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure and conformational flexibility of the unligated FK506-binding protein FKBP12.6.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4IQC
 
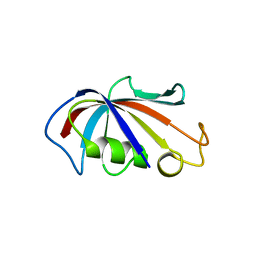 | | P3121 crystal form of FKBP12.6 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B | | Authors: | Chen, H, Mustafi, S.M, Li, H.M, LeMaster, D.M, Hernandez, G. | | Deposit date: | 2013-01-11 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Crystal structure and conformational flexibility of the unligated FK506-binding protein FKBP12.6.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4R0X
 
 | | Allosteric coupling of conformational transitions in the FK1 domain of FKBP51 near the site of steroid receptor interaction | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | LeMaster, D.M, Mustafi, S.M, Brecher, M, Zhang, J, Heroux, A, Li, H.M, Hernandez, G. | | Deposit date: | 2014-08-02 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Coupling of Conformational Transitions in the N-terminal Domain of the 51-kDa FK506-binding Protein (FKBP51) Near Its Site of Interaction with the Steroid Receptor Proteins.
J.Biol.Chem., 290, 2015
|
|
5CUQ
 
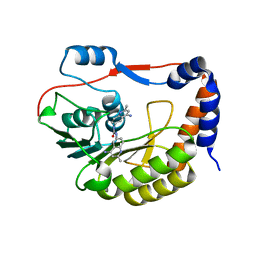 | | Identification and characterization of novel broad spectrum inhibitors of the flavivirus methyltransferase | | Descriptor: | N,N'-BIS(4-AMINO-2-METHYLQUINOLIN-6-YL)UREA, Nonstructural protein NS5 | | Authors: | Brecher, B, Chen, H, Li, Z, Banavali, N.K, Jones, S.A, Zhang, J, Kramer, L.D, Li, H.M. | | Deposit date: | 2015-07-24 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Identification and Characterization of Novel Broad-Spectrum Inhibitors of the Flavivirus Methyltransferase.
Acs Infect Dis., 1, 2015
|
|
4MO7
 
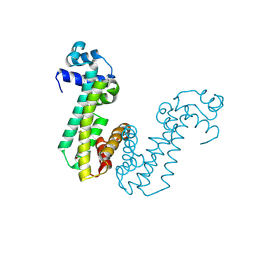 | | Crystal structure of superantigen PfiT | | Descriptor: | BETA-MERCAPTOETHANOL, MAGNESIUM ION, Transcriptional regulator I2 | | Authors: | Liu, L.H, Chen, H, Li, H.M. | | Deposit date: | 2013-09-11 | | Release date: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Pfit is a structurally novel Crohn's disease-associated superantigen.
Plos Pathog., 9, 2013
|
|
4MXM
 
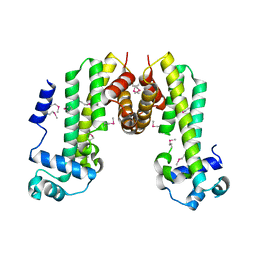 | |
4IPX
 
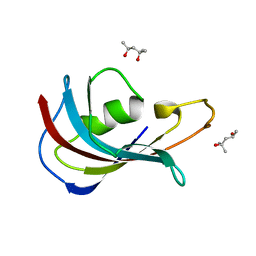 | | Analyzing the visible conformational substates of the FK506 binding protein FKBP12 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Peptidyl-prolyl cis-trans isomerase FKBP1A | | Authors: | Chen, H, Mustafi, S.M, Li, H.M, LeMaster, D.M, Hernandez, G. | | Deposit date: | 2013-01-10 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Analysing the visible conformational substates of the FK506-binding protein FKBP12.
Biochem.J., 453, 2013
|
|
4N19
 
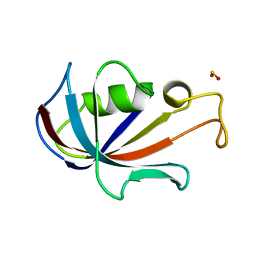 | | Structural basis of conformational transitions in the active site and 80 s loop in the FK506 binding protein FKBP12 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1A, SULFATE ION | | Authors: | Mustafi, S.M, Brecher, M.B, Zhang, J, Li, H.M, Lemaster, D.M, Hernandez, G. | | Deposit date: | 2013-10-03 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural basis of conformational transitions in the active site and 80's loop in the FK506-binding protein FKBP12.
Biochem.J., 458, 2014
|
|
1T7B
 
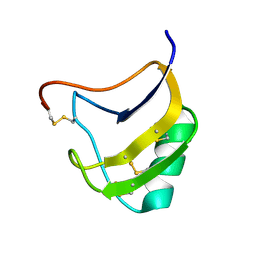 | | Crystal structure of mutant Lys8Gln of scorpion alpha-like neurotoxin BmK M1 from Buthus martensii Karsch | | Descriptor: | Alpha-like neurotoxin BmK-I | | Authors: | Xiang, Y, Guan, R.J, He, X.L, Wang, C.G, Wang, M, Zhang, Y, Sundberg, E.J, Wang, D.C. | | Deposit date: | 2004-05-09 | | Release date: | 2004-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Mechanism Governing Cis and Trans Isomeric States and an Intramolecular Switch for Cis/Trans Isomerization of a Non-proline Peptide Bond Observed in Crystal Structures of Scorpion Toxins
J.Mol.Biol., 341, 2004
|
|
1T7A
 
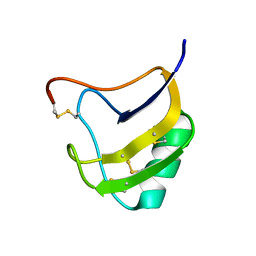 | | Crystal structure of mutant Lys8Asp of scorpion alpha-like neurotoxin BmK M1 from Buthus martensii Karsch | | Descriptor: | Alpha-like neurotoxin BmK-I | | Authors: | Xiang, Y, Guan, R.J, He, X.L, Wang, C.G, Wang, M, Zhang, Y, Sundberg, E.J, Wang, D.C. | | Deposit date: | 2004-05-08 | | Release date: | 2004-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Mechanism Governing Cis and Trans Isomeric States and an Intramolecular Switch for Cis/Trans Isomerization of a Non-proline Peptide Bond Observed in Crystal Structures of Scorpion Toxins
J.Mol.Biol., 341, 2004
|
|
