1IBG
 
 | |
1BTJ
 
 | | HUMAN SERUM TRANSFERRIN, RECOMBINANT N-TERMINAL LOBE, APO FORM, CRYSTAL FORM 2 | | Descriptor: | PROTEIN (SERUM TRANSFERRIN) | | Authors: | Jeffrey, P.D, Bewley, M.C, Macgillivray, R.T.A, Mason, A.B, Woodworth, R.C, Baker, E.N. | | Deposit date: | 1998-09-01 | | Release date: | 1999-01-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Ligand-induced conformational change in transferrins: crystal structure of the open form of the N-terminal half-molecule of human transferrin.
Biochemistry, 37, 1998
|
|
1BP5
 
 | | HUMAN SERUM TRANSFERRIN, RECOMBINANT N-TERMINAL LOBE, APO FORM | | Descriptor: | PROTEIN (SERUM TRANSFERRIN) | | Authors: | Jeffrey, P.D, Bewley, M.C, Macgillivray, R.T.A, Mason, A.B, Woodworth, R.C, Baker, E.N. | | Deposit date: | 1998-08-12 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ligand-induced conformational change in transferrins: crystal structure of the open form of the N-terminal half-molecule of human transferrin.
Biochemistry, 37, 1998
|
|
1C26
 
 | |
1IGJ
 
 | |
1IGI
 
 | |
1FIN
 
 | |
1G3N
 
 | |
7S97
 
 | | Structure of the Photoacclimated Light Harvesting Complex PC577 from Hemiselmis pacifica | | Descriptor: | 15,16-DIHYDROBILIVERDIN, PHYCOCYANOBILIN, Phycoerythrin alpha subunit 1, ... | | Authors: | Jeffrey, P.D, Spangler, L.C, Scholes, G.D. | | Deposit date: | 2021-09-20 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Controllable Phycobilin Modification: An Alternative Photoacclimation Response in Cryptophyte Algae.
Acs Cent.Sci., 8, 2022
|
|
7S96
 
 | | Structure of the Light Harvesting Complex PC577 from Hemiselmis pacifica | | Descriptor: | 15,16-DIHYDROBILIVERDIN, PHYCOCYANOBILIN, Phycoerythrin alpha subunit 1, ... | | Authors: | Jeffrey, P.D, Spangler, L.C, Scholes, G.D. | | Deposit date: | 2021-09-20 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Controllable Phycobilin Modification: An Alternative Photoacclimation Response in Cryptophyte Algae.
Acs Cent.Sci., 8, 2022
|
|
3UO8
 
 | | Crystal structure of the MALT1 paracaspase (P1 form) | | Descriptor: | Mucosa-associated lymphoid tissue lymphoma translocation protein 1, Z-Val-Arg-Pro-DL-Arg-fluoromethylketone | | Authors: | Jeffrey, P.D, Yu, J.W, Shi, Y. | | Deposit date: | 2011-11-16 | | Release date: | 2011-12-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the mucosa-associated lymphoid tissue lymphoma translocation 1 (MALT1) paracaspase region.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3UOA
 
 | | Crystal structure of the MALT1 paracaspase (P21 form) | | Descriptor: | MAGNESIUM ION, Mucosa-associated lymphoid tissue lymphoma translocation protein 1, Z-Val-Arg-Pro-DL-Arg-fluoromethylketone | | Authors: | Jeffrey, P.D, Yu, J.W, Shi, Y. | | Deposit date: | 2011-11-16 | | Release date: | 2011-12-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the mucosa-associated lymphoid tissue lymphoma translocation 1 (MALT1) paracaspase region.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5IT3
 
 | | Swirm domain of human Lsd1 | | Descriptor: | Lysine-specific histone demethylase 1A, MAGNESIUM ION | | Authors: | Jeffrey, P.D, Yuan, P. | | Deposit date: | 2016-03-16 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A Tlx-interacting peptide of Lsd1 inhibits the proliferation of brain tumor stem cells
To Be Published
|
|
3B4R
 
 | | Site-2 Protease from Methanocaldococcus jannaschii | | Descriptor: | Putative zinc metalloprotease MJ0392, ZINC ION | | Authors: | Jeffrey, P.D, Feng, L, Yan, H, Wu, Z, Yan, N, Wang, Z, Shi, Y. | | Deposit date: | 2007-10-24 | | Release date: | 2008-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of a site-2 protease family intramembrane metalloprotease.
Science, 318, 2007
|
|
2P1L
 
 | |
2PKG
 
 | |
6WB4
 
 | | Microbiome-derived Acarbose Kinase Mak1 Labeled with selenomethionine | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, Acarbose Kinase Mak1, ... | | Authors: | Jeffrey, P.D, Balaich, J.N, Estrella, M.A, Donia, M.S. | | Deposit date: | 2020-03-26 | | Release date: | 2021-04-21 | | Last modified: | 2021-12-15 | | Method: | X-RAY DIFFRACTION (2.593 Å) | | Cite: | The human microbiome encodes resistance to the antidiabetic drug acarbose.
Nature, 600, 2021
|
|
6WB5
 
 | | Microbiome-derived Acarbose Kinase Mak1 as a Complex with Acarbose and AMP-PNP | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Acarbose Kinase Mak1, MANGANESE (II) ION, ... | | Authors: | Jeffrey, P.D, Balaich, J.N, Estrella, M.A, Donia, M.S. | | Deposit date: | 2020-03-26 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | The human microbiome encodes resistance to the antidiabetic drug acarbose.
Nature, 600, 2021
|
|
6WB7
 
 | | Acarbose Kinase AcbK as a Complex with Acarbose and AMP-PNP | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Acarbose 7(IV)-phosphotransferase, MANGANESE (II) ION, ... | | Authors: | Jeffrey, P.D, Balaich, J.N, Estrella, M.A, Donia, M.S. | | Deposit date: | 2020-03-26 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | The human microbiome encodes resistance to the antidiabetic drug acarbose.
Nature, 600, 2021
|
|
7TLF
 
 | | Structure of the photoacclimated Light Harvesting Complex PE545 from Proteomonas sulcata | | Descriptor: | 15,16-DIHYDROBILIVERDIN, PHYCOERYTHROBILIN, Phycoerythrin alpha-subunit 1, ... | | Authors: | Jeffrey, P.D, Spangler, L.C, Scholes, G.D. | | Deposit date: | 2022-01-18 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Controllable Phycobilin Modification: An Alternative Photoacclimation Response in Cryptophyte Algae.
Acs Cent.Sci., 8, 2022
|
|
7TJA
 
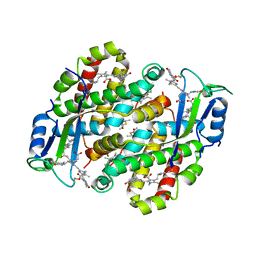 | | Structure of the Light Harvesting Complex PE545 from Proteomonas sulcata | | Descriptor: | 15,16-DIHYDROBILIVERDIN, MAGNESIUM ION, PHYCOERYTHROBILIN, ... | | Authors: | Jeffrey, P.D, Spangler, L.C, Scholes, G.D. | | Deposit date: | 2022-01-15 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Controllable Phycobilin Modification: An Alternative Photoacclimation Response in Cryptophyte Algae.
Acs Cent.Sci., 8, 2022
|
|
7TZ9
 
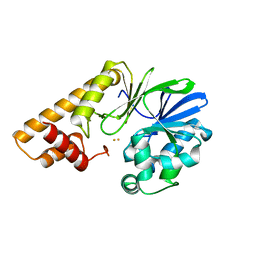 | |
7TZA
 
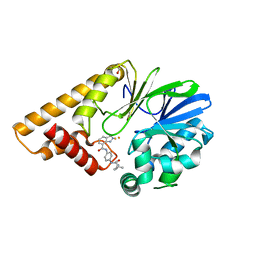 | | Structure of PQS Response Protein PqsE in complex with N-(4-(3-neopentylureido)phenyl)-1H-indazole-7-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, N-{4-[(2,2-dimethylpropyl)carbamamido]phenyl}-1H-indazole-7-carboxamide, ... | | Authors: | Jeffrey, P.D, Taylor, I.R, Bassler, B.L. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The PqsE Active Site as a Target for Small Molecule Antimicrobial Agents against Pseudomonas aeruginosa.
Biochemistry, 61, 2022
|
|
7U6G
 
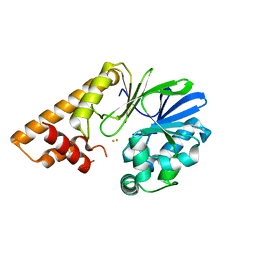 | |
6XJL
 
 | |
