6ELK
 
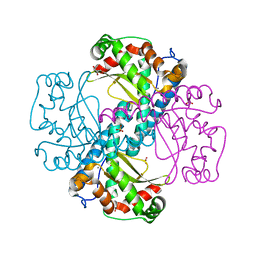 | | C.elegans MnSOD-3 mutant - Q142H | | Descriptor: | GLYCEROL, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Hunter, G.J, Trinh, C.H, Hunter, T. | | Deposit date: | 2017-09-29 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Single Mutation is Sufficient to Modify the Metal Selectivity and Specificity of a Eukaryotic Manganese Superoxide Dismutase to Encompass Iron.
Chemistry, 24, 2018
|
|
4X9Q
 
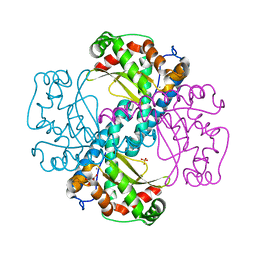 | | MnSOD-3 Room Temperature Structure | | Descriptor: | MALONATE ION, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Hunter, G.J, Trinh, C.H, Hunter, T, Bonetta, R, Stewart, E.E. | | Deposit date: | 2014-12-11 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The structure of the Caenorhabditis elegans manganese superoxide dismutase MnSOD-3-azide complex.
Protein Sci., 24, 2015
|
|
5AG2
 
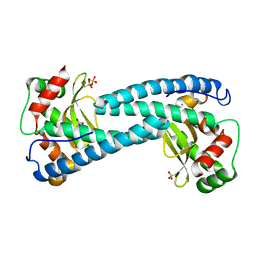 | | SOD-3 azide complex | | Descriptor: | ACETATE ION, AZIDE ION, MALONATE ION, ... | | Authors: | Hunter, G.J, Trinh, C.H, Bonetta, R, Stewart, E.E, Cabelli, D.E, Hunter, T. | | Deposit date: | 2015-01-27 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The Structure of the Caenorhabditis Elegans Manganese Superoxide Dismutase Mnsod-3-Azide Complex.
Protein Sci., 24, 2015
|
|
3DC6
 
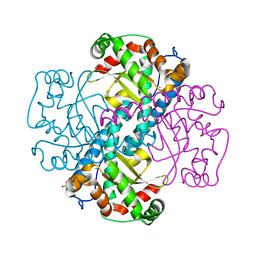 | | Crystal Structure of a manganese superoxide dismutases from Caenorhabditis elegans | | Descriptor: | MANGANESE (II) ION, SULFATE ION, Superoxide dismutase [Mn] 1 | | Authors: | Trinh, C.H, Hunter, T, Stewart, E.E, Phillips, S.E.V, Hunter, G.J. | | Deposit date: | 2008-06-03 | | Release date: | 2009-06-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Purification, crystallization and X-ray structures of the two manganese superoxide dismutases from Caenorhabditis elegans
Acta Crystallogr.,Sect.F, 64, 2008
|
|
3DC5
 
 | | Crystal Structure of a manganese superoxide dismutases from Caenorhabditis elegans | | Descriptor: | MALONATE ION, MANGANESE (II) ION, Superoxide dismutase [Mn] 2 | | Authors: | Trinh, C.H, Hunter, T, Stewart, E.E, Phillips, S.E.V, Hunter, G.J. | | Deposit date: | 2008-06-03 | | Release date: | 2009-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Purification, crystallization and X-ray structures of the two manganese superoxide dismutases from Caenorhabditis elegans
Acta Crystallogr.,Sect.F, 64, 2008
|
|
6QZN
 
 | | H30 MnSOD-3 Mutant III | | Descriptor: | MANGANESE (II) ION, SULFATE ION, Superoxide dismutase [Mn] 2, ... | | Authors: | Bonetta, R, Trinh, C.H, Hunter, G.J, Hunter, T. | | Deposit date: | 2019-03-11 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | H30 MnSOD-3 Mutant II
To Be Published
|
|
6QZM
 
 | | H30 MnSOD-3 Mutant I | | Descriptor: | MANGANESE (II) ION, SULFATE ION, Superoxide dismutase [Mn] 2, ... | | Authors: | Bonetta, R, Trinh, C.H, Hunter, G.J, Hunter, T. | | Deposit date: | 2019-03-11 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | H30 MnSOD-3 Mutant II
To Be Published
|
|
6S0D
 
 | | H30 MnSOD-3 Mutant II | | Descriptor: | MANGANESE (II) ION, SULFATE ION, Superoxide dismutase [Mn] 2, ... | | Authors: | Bonetta, R, Trinh, C.H, Hunter, G.J, Hunter, T. | | Deposit date: | 2019-06-14 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | H30 MnSOD-3 Mutant II
To Be Published
|
|
6X1V
 
 | | Structure of pHis Fab (SC44-8) in complex with pHis mimetic peptide | | Descriptor: | 1,2-ETHANEDIOL, ACLYana-3-pTza peptide, SC44-8 Heavy chain, ... | | Authors: | Kalagiri, R, Stanfield, R, Wilson, I.A, Hunter, T. | | Deposit date: | 2020-05-19 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural basis for differential recognition of phosphohistidine-containing peptides by 1-pHis and 3-pHis monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6X1T
 
 | | Structure of pHis Fab (SC50-3) in complex with pHis mimetic peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kalagiri, R, Stanfield, R.L, Wilson, I.A, Hunter, T. | | Deposit date: | 2020-05-19 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural basis for differential recognition of phosphohistidine-containing peptides by 1-pHis and 3-pHis monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6X1W
 
 | | Structure of pHis Fab (SC56-2) in complex with pHis mimetic peptide | | Descriptor: | ACLYana-3-pTza peptide, SC56-2 Heavy chain, SC56-2 Light chain | | Authors: | Kalagiri, R, Stanfield, R, Wilson, I.A, Hunter, T. | | Deposit date: | 2020-05-19 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for differential recognition of phosphohistidine-containing peptides by 1-pHis and 3-pHis monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6X1U
 
 | | Structure of pHis Fab (SC39-4) in complex with pHis mimetic peptide | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ACLYana-3-pTza peptide, ... | | Authors: | Kalagiri, R, Stanfield, R, Wilson, I.A, Hunter, T. | | Deposit date: | 2020-05-19 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.641 Å) | | Cite: | Structural basis for differential recognition of phosphohistidine-containing peptides by 1-pHis and 3-pHis monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6X1S
 
 | | Structure of pHis Fab (SC1-1) in complex with pHis mimetic peptide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, NM23-1-pTza peptide, SC1-1 Heavy chain, ... | | Authors: | Kalagiri, R, Stanfield, R.L, Wilson, I.A, Hunter, T. | | Deposit date: | 2020-05-19 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for differential recognition of phosphohistidine-containing peptides by 1-pHis and 3-pHis monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1PIN
 
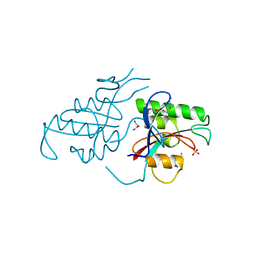 | | PIN1 PEPTIDYL-PROLYL CIS-TRANS ISOMERASE FROM HOMO SAPIENS | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, ALANINE, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE, ... | | Authors: | Noel, J.P, Ranganathan, R, Hunter, T. | | Deposit date: | 1998-06-21 | | Release date: | 1998-10-14 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural and functional analysis of the mitotic rotamase Pin1 suggests substrate recognition is phosphorylation dependent.
Cell(Cambridge,Mass.), 89, 1997
|
|
1F3H
 
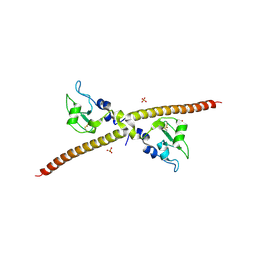 | | X-RAY CRYSTAL STRUCTURE OF THE HUMAN ANTI-APOPTOTIC PROTEIN SURVIVIN | | Descriptor: | SULFATE ION, SURVIVIN, ZINC ION | | Authors: | Verdecia, M.A, Huang, H, Dutil, E, Hunter, T, Noel, J.P. | | Deposit date: | 2000-06-03 | | Release date: | 2000-12-06 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structure of the human anti-apoptotic protein survivin reveals a dimeric arrangement.
Nat.Struct.Biol., 7, 2000
|
|
1F8A
 
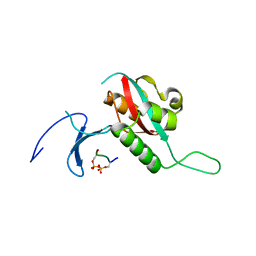 | | STRUCTURAL BASIS FOR THE PHOSPHOSERINE-PROLINE RECOGNITION BY GROUP IV WW DOMAINS | | Descriptor: | PEPTIDYL-PROLYL CIS-TRANS ISOMERASE NIMA-INTERACTING 1, Y(SEP)PT(SEP)S PEPTIDE | | Authors: | Verdecia, M.A, Bowman, M.E, Lu, K.P, Hunter, T, Noel, J.P. | | Deposit date: | 2000-06-29 | | Release date: | 2000-08-23 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural basis for phosphoserine-proline recognition by group IV WW domains.
Nat.Struct.Biol., 7, 2000
|
|
1ND7
 
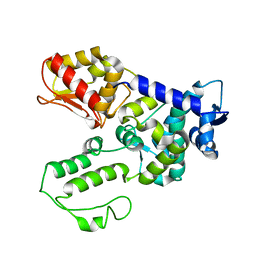 | | Conformational Flexibility Underlies Ubiquitin Ligation Mediated by the WWP1 HECT domain E3 Ligase | | Descriptor: | WW domain-containing protein 1 | | Authors: | Verdecia, M.A, Joaziero, C.A.P, Wells, N.J, Ferrer, J.-L, Bowman, M.E, Hunter, T, Noel, J.P. | | Deposit date: | 2002-12-08 | | Release date: | 2003-09-23 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational Flexibility Underlies Ubiquitin Ligation Mediated by the WWP1 HECT domain E3 Ligase
Mol.Cell, 11, 2003
|
|
1P15
 
 | |
1P13
 
 | | Crystal Structure of the Src SH2 Domain Complexed with Peptide (SDpYANFK) | | Descriptor: | CACODYLATE ION, Peptide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Sonnenburg, E.D, Bilwes, A, Hunter, T, Noel, J.P. | | Deposit date: | 2003-04-11 | | Release date: | 2003-08-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | The structure of the membrane distal phosphatase domain of RPTPalpha reveals interdomain flexibility and an SH2 domain interaction region.
Biochemistry, 42, 2003
|
|
4UY9
 
 | | Structure of MLK1 kinase domain with leucine zipper 1 | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 9 | | Authors: | Read, J.A, Brassington, C, Pollard, H.K, Phillips, C, Green, I, Overmann, R, Collier, M. | | Deposit date: | 2014-08-29 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Recurrent Mlk4 Loss-of-Function Mutations Suppress Jnk Signaling to Promote Colon Tumorigenesis.
Cancer Res., 76, 2016
|
|
4UYA
 
 | | Structure of MLK4 kinase domain with ATPgammaS | | Descriptor: | MAGNESIUM ION, MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE MLK4, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Read, J.A, Brassington, C, Pollard, H.K, Phillips, C, Green, I, Overmann, R, Collier, M. | | Deposit date: | 2014-08-29 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Recurrent Mlk4 Loss-of-Function Mutations Suppress Jnk Signaling to Promote Colon Tumorigenesis.
Cancer Res., 76, 2016
|
|
3NG2
 
 | |
1NMW
 
 | | Solution structure of the PPIase domain of human Pin1 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, SULFATE ION | | Authors: | Bayer, E, Goettsch, S, Mueller, J.W, Griewel, B, Guiberman, E, Mayr, L, Bayer, P. | | Deposit date: | 2003-01-12 | | Release date: | 2003-07-15 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of the Mitotic Regulator hPin1 in Solution: INSIGHTS INTO DOMAIN ARCHITECTURE AND SUBSTRATE BINDING.
J.Biol.Chem., 278, 2003
|
|
1NMV
 
 | | Solution structure of human Pin1 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Bayer, E, Goettsch, S, Mueller, J.W, Griewel, B, Guiberman, E, Mayr, L, Bayer, P. | | Deposit date: | 2003-01-11 | | Release date: | 2003-08-12 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of the Mitotic Regulator hPin1 in Solution: INSIGHTS INTO DOMAIN ARCHITECTURE AND SUBSTRATE BINDING.
J.Biol.Chem., 278, 2003
|
|
2Q5A
 
 | | human Pin1 bound to L-PEPTIDE | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE, Five residue peptide, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Noel, J.P, Zhang, Y. | | Deposit date: | 2007-05-31 | | Release date: | 2007-06-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for high-affinity peptide inhibition of human Pin1.
Acs Chem.Biol., 2, 2007
|
|
