1JLI
 
 | |
6SXN
 
 | | Crystal structure of P212121 apo form of CrtE | | Descriptor: | Geranylgeranyl pyrophosphate synthase | | Authors: | Feng, Y, Morgan, R.M.L, Nixon, P.J. | | Deposit date: | 2019-09-26 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Crystal Structure of Geranylgeranyl Pyrophosphate Synthase (CrtE) Involved in Cyanobacterial Terpenoid Biosynthesis.
Front Plant Sci, 11, 2020
|
|
6SXL
 
 | | Crystal structure of CrtE | | Descriptor: | Geranylgeranyl pyrophosphate synthase, PHOSPHATE ION | | Authors: | Feng, Y, Morgan, R.M.L, Nixon, P.J. | | Deposit date: | 2019-09-26 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Geranylgeranyl Pyrophosphate Synthase (CrtE) Involved in Cyanobacterial Terpenoid Biosynthesis.
Front Plant Sci, 11, 2020
|
|
4ZH2
 
 | |
4ZH3
 
 | |
4ZH4
 
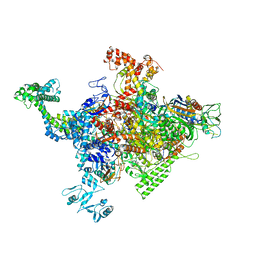
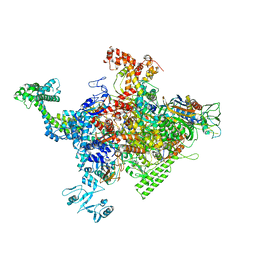 | | Crystal structure of Escherichia coli RNA polymerase in complex with CBRP18 | | Descriptor: | 5-(4-fluorophenyl)-4-[4-fluoro-3-(trifluoromethyl)phenyl]-1H-pyrazole, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Feng, Y, Ebright, R.H. | | Deposit date: | 2015-04-24 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.993 Å) | | Cite: | Structural Basis of Transcription Inhibition by CBR Hydroxamidines and CBR Pyrazoles.
Structure, 23, 2015
|
|
1HOV
 
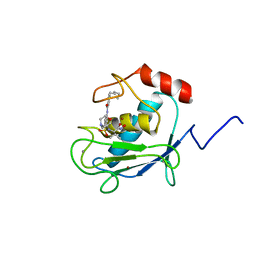 | | SOLUTION STRUCTURE OF A CATALYTIC DOMAIN OF MMP-2 COMPLEXED WITH SC-74020 | | Descriptor: | CALCIUM ION, MATRIX METALLOPROTEINASE-2, N-{4-[(1-HYDROXYCARBAMOYL-2-METHYL-PROPYL)-(2-MORPHOLIN-4-YL-ETHYL)-SULFAMOYL]-4-PENTYL-BENZAMIDE, ... | | Authors: | Feng, Y, Likos, J.J, Zhu, L, Woodward, H, Munie, G, McDonald, J.J, Stevens, A.M, Howard, C.P, De Crescenzo, G.A, Welsch, D, Shieh, H.-S, Stallings, W.C. | | Deposit date: | 2000-12-11 | | Release date: | 2001-12-12 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of the catalytic domain of matrix metalloproteinase-2 complexed with a hydroxamic acid inhibitor
Biochim.Biophys.Acta, 1598, 2002
|
|
2LEO
 
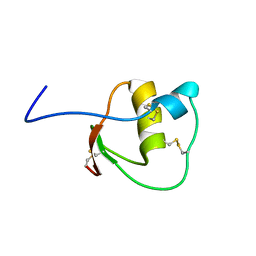 | |
1Z7R
 
 | | Solution Structure of reduced glutaredoxin C1 from Populus tremula x tremuloides | | Descriptor: | glutaredoxin | | Authors: | Feng, Y, Zhong, N, Rouhier, N, Jacquot, J.P, Xia, B. | | Deposit date: | 2005-03-26 | | Release date: | 2006-03-28 | | Last modified: | 2020-02-26 | | Method: | SOLUTION NMR | | Cite: | Structural Insight into Poplar Glutaredoxin C1 with a Bridging Iron-Sulfur Cluster at the Active Site
Biochemistry, 45, 2006
|
|
1Z7P
 
 | | Solution structure of reduced glutaredoxin C1 from Populus tremula x tremuloides | | Descriptor: | glutaredoxin | | Authors: | Feng, Y, Zhong, N, Rouhier, N, Jacquot, J.P, Xia, B. | | Deposit date: | 2005-03-26 | | Release date: | 2006-03-28 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structural Insight into Poplar Glutaredoxin C1 with a Bridging Iron-Sulfur Cluster at the Active Site
Biochemistry, 45, 2006
|
|
3CVA
 
 | | Human Bcl-xL containing a Trp to Ala mutation at position 137 | | Descriptor: | Apoptosis regulator Bcl-X | | Authors: | Feng, Y, Zhang, L, Hu, T, Shen, X, Chen, K, Jiang, H, Liu, D. | | Deposit date: | 2008-04-18 | | Release date: | 2009-03-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A conserved hydrophobic core at Bcl-x(L) mediates its structural stability and binding affinity with BH3-domain peptide of pro-apoptotic protein
Arch.Biochem.Biophys., 484, 2009
|
|
3KXT
 
 | | Crystal structure of Sulfolobus Cren7-dsDNA complex | | Descriptor: | 5'-D(*GP*CP*GP*AP*TP*CP*GP*C)-3', Chromatin protein Cren7 | | Authors: | Feng, Y, Wang, J. | | Deposit date: | 2009-12-03 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Crystal structure of the crenarchaeal conserved chromatin protein Cren7 and double-stranded DNA complex
Protein Sci., 19, 2010
|
|
2HR9
 
 | |
2KJG
 
 | |
5I2D
 
 | | Crystal structure of T. thermophilus TTHB099 class II transcription activation complex: TAP-RPo | | Descriptor: | DNA (72-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Feng, Y, Zhang, Y, Ebright, R.H. | | Deposit date: | 2016-02-08 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4.405 Å) | | Cite: | Structural basis of transcription activation.
Science, 352, 2016
|
|
2MMP
 
 | | Solution structure of a ribosomal protein | | Descriptor: | Uncharacterized protein | | Authors: | Feng, Y. | | Deposit date: | 2014-03-17 | | Release date: | 2014-06-11 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure determination of archaea-specific ribosomal protein L46a reveals a novel protein fold.
Biochem.Biophys.Res.Commun., 450, 2014
|
|
7VRS
 
 | | The complex of Acyltransferase and Acyl Carrier Protein Domains from module 9 of Salinomycin Polyketide Synthase | | Descriptor: | 1,1'-butane-1,4-diylbis(1H-pyrrole-2,5-dione), 4'-PHOSPHOPANTETHEINE, Type I modular polyketide synthase | | Authors: | Feng, Y, Zheng, J. | | Deposit date: | 2021-10-24 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural visualization of transient interactions between the cis-acting acyltransferase and acyl carrier protein of the salinomycin modular polyketide synthase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7VWK
 
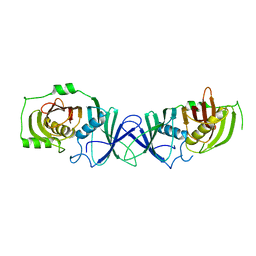 | | The product template domain of AviM | | Descriptor: | Polyketide synthase | | Authors: | Feng, Y, Yang, X, Zheng, J. | | Deposit date: | 2021-11-10 | | Release date: | 2022-06-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Streptomyces viridochromogenes product template domain represents an evolutionary intermediate between dehydratase and aldol cyclase of type I polyketide synthases.
Commun Biol, 5, 2022
|
|
7VT1
 
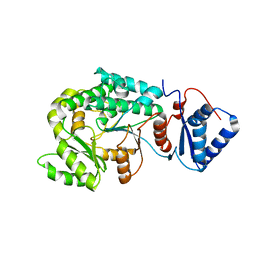 | |
5H9U
 
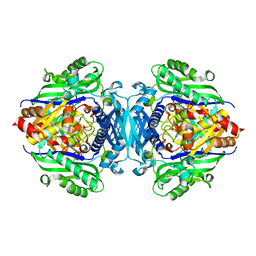 | |
8ILT
 
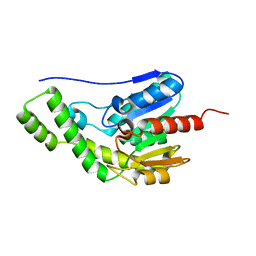 | | Crystal structure of Est30 | | Descriptor: | Carboxylesterase | | Authors: | Feng, Y, Luo, Z. | | Deposit date: | 2023-03-04 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Crystal structure of Est30
To Be Published
|
|
6IGM
 
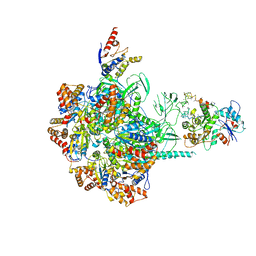 | | Cryo-EM Structure of Human SRCAP Complex | | Descriptor: | Actin-related protein 6, Helicase SRCAP, RuvB-like 1, ... | | Authors: | Feng, Y, Tian, Y, Wu, Z, Xu, Y. | | Deposit date: | 2018-09-25 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of human SRCAP complex.
Cell Res., 28, 2018
|
|
6LP2
 
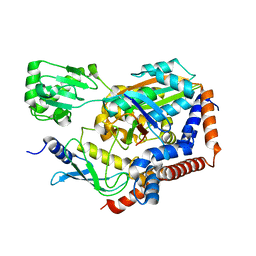 | | Structure of Lpg2148/UBE2N-Ub complex | | Descriptor: | Ubiquitin, Ubiquitin-conjugating enzyme E2 N, Uncharacterized protein lpg2148 | | Authors: | Feng, Y, Wang, Y, Huang, Y, Li, D. | | Deposit date: | 2020-01-08 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.479 Å) | | Cite: | Structure of Lpg2148/UBE2N-Ub complex
To Be Published
|
|
6LW4
 
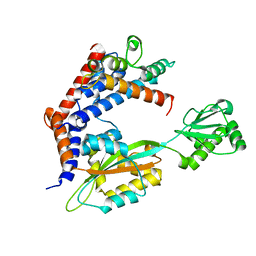 | |
8IWH
 
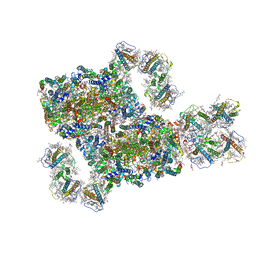 | | Structure and characteristics of a photosystem II supercomplex containing monomeric LHCX and dimeric FCPII antennae from the diatom Thalassiosira pseudonana | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15-octaen-17-ynyl]cyclohex-3-en-1-ol, (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, ... | | Authors: | Feng, Y, Li, Z.H, Wang, W.D, Shen, J.R. | | Deposit date: | 2023-03-30 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structure of a diatom photosystem II supercomplex containing a member of Lhcx family and dimeric FCPII.
Sci Adv, 9, 2023
|
|
