1UCT
 
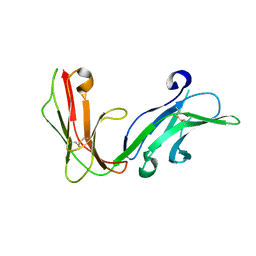 | | Crystal structure of the extracellular fragment of Fc alpha Receptor I (CD89) | | Descriptor: | Immunoglobulin alpha Fc receptor | | Authors: | Ding, Y, Xu, G, Yang, M, Zhang, W, Rao, Z. | | Deposit date: | 2003-04-21 | | Release date: | 2003-07-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Ectodomain of Human Fc{alpha}RI.
J.Biol.Chem., 278, 2003
|
|
8IA7
 
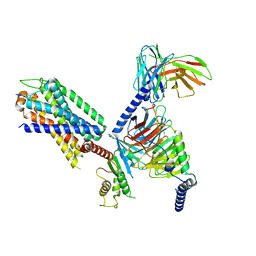 | | Structural insights into human brain gut peptide cholecystokinin receptors | | Descriptor: | CCK-8, Gastrin/cholecystokinin type B receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ding, Y, Zhang, H, Liao, Y, Chen, L, Ji, S. | | Deposit date: | 2023-02-08 | | Release date: | 2023-12-06 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into human brain-gut peptide cholecystokinin receptors.
Cell Discov, 8, 2022
|
|
1J4G
 
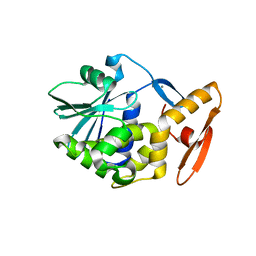 | | crystal structure analysis of the trichosanthin delta C7 | | Descriptor: | CALCIUM ION, Trichosanthin delta C7 | | Authors: | Ding, Y, Too, H.M, Wang, Z.L, Liu, Y.W, Dong, Y.C, Shaw, P.C, Rao, Z.H. | | Deposit date: | 2001-09-30 | | Release date: | 2001-10-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | crystal structure analysis of the trichosanthin delta C7
To be Published
|
|
8Y9S
 
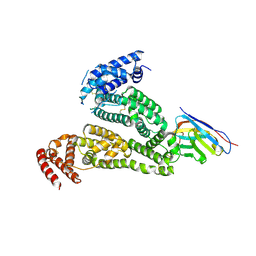 | |
8Y9T
 
 | |
8Y9U
 
 | |
7XOU
 
 | | Structural insights into human brain gut peptide cholecystokinin receptors | | Descriptor: | CCK-8, Cholecystokinin receptor type A, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ding, Y, Zhang, H, Liao, Y, Chen, L, Ji, S. | | Deposit date: | 2022-05-01 | | Release date: | 2022-07-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into human brain-gut peptide cholecystokinin receptors.
Cell Discov, 8, 2022
|
|
7XOV
 
 | | Structural insights into human brain gut peptide cholecystokinin receptors | | Descriptor: | 2-[2-[[4-(4-chloranyl-2,5-dimethoxy-phenyl)-5-(2-cyclohexylethyl)-1,3-thiazol-2-yl]carbamoyl]-5,7-dimethyl-indol-1-yl]ethanoic acid, Cholecystokinin receptor type A, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ding, Y, Zhang, H, Liao, Y, Chen, L, Ji, S. | | Deposit date: | 2022-05-01 | | Release date: | 2022-07-20 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into human brain-gut peptide cholecystokinin receptors.
Cell Discov, 8, 2022
|
|
7XOW
 
 | | Structural insights into human brain gut peptide cholecystokinin receptors | | Descriptor: | Gastrin, Gastrin/cholecystokinin type B receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ding, Y, Zhang, H, Liao, Y, Chen, L, Ji, S. | | Deposit date: | 2022-05-01 | | Release date: | 2022-07-20 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into human brain-gut peptide cholecystokinin receptors.
Cell Discov, 8, 2022
|
|
1MI8
 
 | | 2.0 Angstrom crystal structure of a DnaB intein from Synechocystis sp. PCC 6803 | | Descriptor: | DnaB intein | | Authors: | Ding, Y, Chen, X, Ferrandon, S, Xu, M, Rao, Z. | | Deposit date: | 2002-08-22 | | Release date: | 2003-08-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of mini-intein reveals a conserved catalytic module involved in side chain cyclization of asparagine during protein splicing
J.Biol.Chem., 278, 2003
|
|
8K4Q
 
 | |
8J1I
 
 | | Crystal Structure of EphA8/SASH1 Complex | | Descriptor: | Ephrin type-A receptor 8, SAM and SH3 domain-containing protein 1 | | Authors: | Liu, W, Li, J, Ding, Y. | | Deposit date: | 2023-04-12 | | Release date: | 2024-02-28 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | SASH1: A Novel Eph Receptor Partner and Insights into SAM-SAM Interactions.
J.Mol.Biol., 435, 2023
|
|
5ZBZ
 
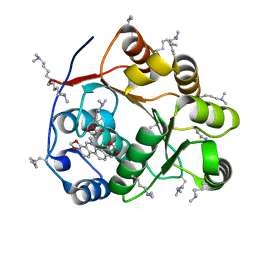 | | Crystal structure of the DEAD domain of Human eIF4A with sanguinarine | | Descriptor: | 13-methyl[1,3]benzodioxolo[5,6-c][1,3]dioxolo[4,5-i]phenanthridin-13-ium, Eukaryotic initiation factor 4A-I, MALONATE ION | | Authors: | Ding, Y, Ding, L. | | Deposit date: | 2018-02-14 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.30860257 Å) | | Cite: | Targeting the N Terminus of eIF4AI for Inhibition of Its Catalytic Recycling.
Cell Chem Biol, 26, 2019
|
|
8JMR
 
 | | Crystal structure of hinokiresinol synthase in complex with 1,7-bis(4-hydroxyphenyl)hepta-1,6-dien-3-one | | Descriptor: | 1,7-bis(4-hydroxyphenyl)hepta-1,6-dien-3-one, Hinokiresinol synthase alpha subunit, Hinokiresinol synthase beta subunit, ... | | Authors: | Ding, Y, Ushimaru, R, Mori, T, Abe, I. | | Deposit date: | 2023-06-05 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Mechanistic Insights into the C-C Bond-Forming Rearrangement Reaction Catalyzed by Heterodimeric Hinokiresinol Synthase.
J.Am.Chem.Soc., 145, 2023
|
|
8JMQ
 
 | | Crystal structure of hinokiresinol synthase | | Descriptor: | Hinokiresinol synthase alpha subunit, Hinokiresinol synthase beta subunit | | Authors: | Ding, Y, Ushimaru, R, Mori, T, Abe, I. | | Deposit date: | 2023-06-05 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Mechanistic Insights into the C-C Bond-Forming Rearrangement Reaction Catalyzed by Heterodimeric Hinokiresinol Synthase.
J.Am.Chem.Soc., 145, 2023
|
|
1GHD
 
 | | Crystal structure of the glutaryl-7-aminocephalosporanic acid acylase by mad phasing | | Descriptor: | GLUTARYL-7-AMINOCEPHALOSPORANIC ACID ACYLASE | | Authors: | Ding, Y, Jiang, W, Mao, X, He, H, Zhang, S, Tang, H, Bartlam, M, Ye, S, Jiang, F, Liu, Y, Zhao, G, Rao, Z. | | Deposit date: | 2000-12-07 | | Release date: | 2003-07-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Affinity alkylation of the Trp-B4 residue of the beta -subunit of the glutaryl 7-aminocephalosporanic acid acylase of Pseudomonas sp. 130.
J.Biol.Chem., 277, 2002
|
|
6ITQ
 
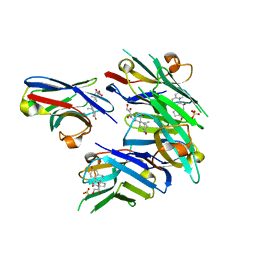 | | Crystal structure of cortisol complexed with its nanobody at pH 10.5 | | Descriptor: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, SULFATE ION, anti-cortisol camelid antibody | | Authors: | Ding, Y, Ding, L.L, Wang, Z.Y, Zhong, P.Y. | | Deposit date: | 2018-11-24 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.526 Å) | | Cite: | Structural insights into the mechanism of single domain VHH antibody binding to cortisol.
Febs Lett., 593, 2019
|
|
6J04
 
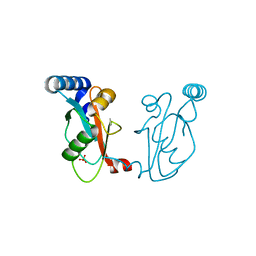 | |
6ITP
 
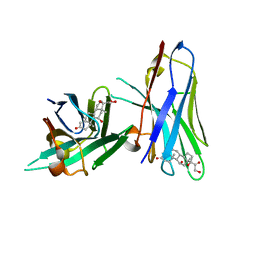 | | Crystal structure of cortisol complexed with its nanobody at pH 3.5 | | Descriptor: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, anti-cortisol camelid antibody | | Authors: | Ding, Y, Ding, L.L, Wang, Z.Y, Zhong, P.Y. | | Deposit date: | 2018-11-24 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.572 Å) | | Cite: | Structural insights into the mechanism of single domain VHH antibody binding to cortisol.
Febs Lett., 593, 2019
|
|
8YBN
 
 | | Crystal structure of nanobody SEB-Nb8 bound to staphylococcal enterotoxin B (SEB) | | Descriptor: | Enterotoxin type B, nanobody SEB-Nb8 | | Authors: | Ding, Y, Zong, X, Liu, R, Liu, P. | | Deposit date: | 2024-02-15 | | Release date: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural insights into the binding of nanobodies to the Staphylococcal enterotoxin B.
Int.J.Biol.Macromol., 276, 2024
|
|
8YBO
 
 | | Crystal structure of nanobody SEB-Nb11 bound to staphylococcal enterotoxin B (SEB) | | Descriptor: | Enterotoxin type B, nanobody SEB-Nb11 | | Authors: | Ding, Y, Zong, X, Liu, R, Liu, P. | | Deposit date: | 2024-02-15 | | Release date: | 2024-12-25 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural insights into the binding of nanobodies to the Staphylococcal enterotoxin B.
Int.J.Biol.Macromol., 276, 2024
|
|
8YBM
 
 | | Crystal structure of nanobody SEB-Nb6 bound to staphylococcal enterotoxin B (SEB) | | Descriptor: | Enterotoxin type B, nanobody SEB-Nb6 | | Authors: | Ding, Y, Zong, X, Liu, R, Liu, P. | | Deposit date: | 2024-02-15 | | Release date: | 2024-12-25 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural insights into the binding of nanobodies to the Staphylococcal enterotoxin B.
Int.J.Biol.Macromol., 276, 2024
|
|
8YBL
 
 | | Crystal structure of nanobody SEB-Nb3 bound to staphylococcal enterotoxin B (SEB) | | Descriptor: | Enterotoxin type B, nanobody SEB-Nb3 | | Authors: | Ding, Y, Zong, X, Liu, R, Liu, P. | | Deposit date: | 2024-02-15 | | Release date: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural insights into the binding of nanobodies to the Staphylococcal enterotoxin B.
Int.J.Biol.Macromol., 276, 2024
|
|
8YBP
 
 | | Crystal structure of nanobody SEB-Nb20 bound to staphylococcal enterotoxin B (SEB) | | Descriptor: | Enterotoxin type B, nanobody SEB-Nb20 | | Authors: | Ding, Y, Zong, X, Liu, R, Liu, P. | | Deposit date: | 2024-02-15 | | Release date: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural insights into the binding of nanobodies to the Staphylococcal enterotoxin B.
Int.J.Biol.Macromol., 276, 2024
|
|
4Z3D
 
 | |
