139D
 
 | |
134D
 
 | |
1HWV
 
 | | MOLECULAR TOPOLOGY OF POLYCYCLIC AROMATIC CARCINOGENS DETERMINES DNA ADDUCT CONFORMATION: A LINK TO TUMORIGENIC ACTIVITY | | Descriptor: | (1S)-1,2,3,4-TETRAHYDRO-BENZO[C]PHENANTHRENE-2,3,4-TRIOL, 5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3' | | Authors: | Patel, D.J, Lin, C.H, Geacintov, N.E, Broyde, S, Huang, X, Kolbanovskii, A, Hingerty, B.E, Amin, S. | | Deposit date: | 2001-01-10 | | Release date: | 2001-03-21 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Molecular topology of polycyclic aromatic carcinogens determines DNA adduct conformation: a link to tumorigenic activity.
J.Mol.Biol., 306, 2001
|
|
1HX4
 
 | | MOLECULAR TOPOLOGY OF POLYCYCLIC AROMATIC CARCINOGENS DETERMINES DNA ADDUCT CONFORMATION: A LINK TO TUMORIGENIC ACTIVITY | | Descriptor: | (1R)-1,2,3,4-TETRAHYDRO-BENZO[C]PHENANTHRENE-2,3,4-TRIOL, 5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3' | | Authors: | Patel, D.J, Lin, C.H, Geacintov, N.E, Broyde, S, Huang, X, Kolbanovskii, A, Hingerty, B.E, Amin, S. | | Deposit date: | 2001-01-11 | | Release date: | 2001-03-21 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Molecular topology of polycyclic aromatic carcinogens determines DNA adduct conformation: a link to tumorigenic activity.
J.Mol.Biol., 306, 2001
|
|
4IUR
 
 | |
149D
 
 | |
1C0Y
 
 | | SOLUTION STRUCTURE OF THE [AF]-C8-DG ADDUCT POSITIONED OPPOSITE DA AT A TEMPLATE-PRIMER JUNCTION | | Descriptor: | 2-AMINOFLUORENE, DNA (5'-D(*AP*AP*CP*GP*CP*TP*AP*CP*CP*AP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*GP*GP*TP*AP*GP*C)-3') | | Authors: | Gu, Z, Gorin, A, Hingerty, B.E, Broyde, S, Patel, D.J. | | Deposit date: | 1999-07-19 | | Release date: | 1999-08-31 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structures of aminofluorene [AF]-stacked conformers of the syn [AF]-C8-dG adduct positioned opposite dC or dA at a template-primer junction.
Biochemistry, 38, 1999
|
|
1AXO
 
 | | STRUCTURAL ALIGNMENT OF THE (+)-TRANS-ANTI-[BP]DG ADDUCT POSITIONED OPPOSITE DC AT A DNA TEMPLATE-PRIMER JUNCTION, NMR, 6 STRUCTURES | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, DNA DUPLEX D(AAC-[BP]G-CTACCATCC)D(GGATGGTAGC) | | Authors: | Feng, B, Gorin, A.A, Hingerty, B.E, Geacintov, N.E, Broyde, S, Patel, D.J. | | Deposit date: | 1997-10-16 | | Release date: | 1998-07-01 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structural alignment of the (+)-trans-anti-benzo[a]pyrene-dG adduct positioned opposite dC at a DNA template-primer junction.
Biochemistry, 36, 1997
|
|
1B6Y
 
 | | 3,N4-ETHENO-2'-DEOXYCYTIDINE OPPOSITE ADENINE IN AN 11-MER DUPLEX, SOLUTION STRUCTURE FROM NMR AND MOLECULAR DYNAMICS, 2 STRUCTURES | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*(EDC)P*CP*AP*TP*GP*C)-3', 5'-D(*GP*CP*AP*TP*GP*AP*GP*TP*AP*CP*G)-3' | | Authors: | Korobka, A, Cullinan, D, Cosman, M, Grollman, A.P, Patel, D.J, Eisenberg, M, De Los Santos, C. | | Deposit date: | 1999-01-19 | | Release date: | 1999-01-27 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an oligodeoxynucleotide duplex containing the exocyclic lesion 3,N4-etheno-2'-deoxycytidine opposite 2'-deoxyadenosine, determined by NMR spectroscopy and restrained molecular dynamics.
Biochemistry, 35, 1996
|
|
1B5K
 
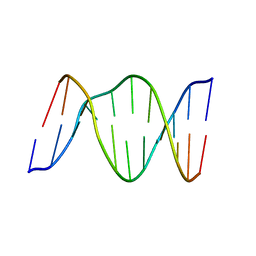 | | 3,N4-ETHENO-2'-DEOXYCYTIDINE OPPOSITE THYMIDINE IN AN 11-MER DUPLEX, SOLUTION STRUCTURE FROM NMR AND MOLECULAR DYNAMICS | | Descriptor: | DNA (5'-D(*CP*GP*TP*AP*CP*EDCP*CP*AP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*TP*GP*TP*AP*CP*G)-3') | | Authors: | Cullinan, D, Korobka, A, Grollman, A.P, Patel, D.J, Eisenberg, M, De Santos, C.L. | | Deposit date: | 1999-01-07 | | Release date: | 1999-01-13 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of an oligodeoxynucleotide duplex containing the exocyclic lesion 3,N4-etheno-2'-deoxycytidine opposite thymidine: comparison with the duplex containing deoxyadenosine opposite the adduct.
Biochemistry, 35, 1996
|
|
1AW4
 
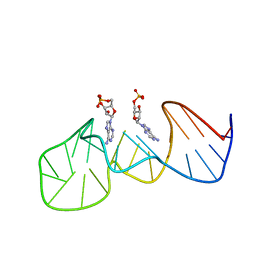 | |
1AXV
 
 | | SOLUTION NMR STRUCTURE OF THE [BP]DA ADDUCT OPPOSITE DT IN A DNA DUPLEX, 6 STRUCTURES | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, DNA DUPLEX D(CTCTC-[BP]A-CTTCC)D(GGAAGTGAGAG) | | Authors: | Mao, B, Gu, Z, Gorin, A.A, Chen, J, Hingerty, B.E, Amid, S, Broyde, S, Geacintov, N.E, Patel, D.J. | | Deposit date: | 1997-10-21 | | Release date: | 1998-07-01 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the (+)-cis-anti-benzo[a]pyrene-dA ([BP]dA) adduct opposite dT in a DNA duplex.
Biochemistry, 38, 1999
|
|
1AX7
 
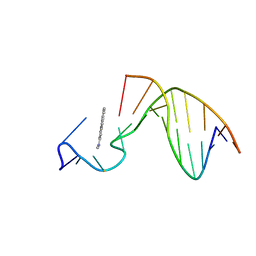 | | SOLUTION STRUCTURE OF THE [AF]-C8-DG ADDUCT POSITIONED AT A TEMPLATE-PRIMER JUNCTION, NMR, 6 STRUCTURES | | Descriptor: | 2-AMINOFLUORENE, DNA DUPLEX D(AAC-[AF]G-CTACCATCC)D(GGATGGTAG) | | Authors: | Mao, B, Gu, Z, Gorin, A.A, Hingerty, B.E, Broyde, S, Patel, D.J. | | Deposit date: | 1997-10-30 | | Release date: | 1998-07-01 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the aminofluorene-stacked conformer of the syn [AF]-C8-dG adduct positioned at a template-primer junction.
Biochemistry, 36, 1997
|
|
1AXU
 
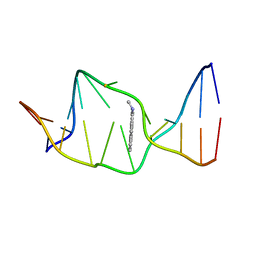 | | SOLUTION NMR STRUCTURE OF THE [AP]DG ADDUCT OPPOSITE DA IN A DNA DUPLEX, NMR, 9 STRUCTURES | | Descriptor: | DNA DUPLEX D(CCATC-[AP]G-CTACC)D(GGTAGAGATGG), N-1-AMINOPYRENE | | Authors: | Gu, Z, Gorin, A.A, Krishnasami, R, Hingerty, B.E, Basu, A.K, Broyde, S, Patel, D.J. | | Deposit date: | 1997-10-21 | | Release date: | 1998-07-01 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-(deoxyguanosin-8-yl)-1-aminopyrene ([AP]dG) adduct opposite dA in a DNA duplex.
Biochemistry, 38, 1999
|
|
1BIV
 
 | | BOVINE IMMUNODEFICIENCY VIRUS TAT-TAR COMPLEX, NMR, 5 STRUCTURES | | Descriptor: | TAR RNA, TAT PEPTIDE | | Authors: | Ye, X, Kumar, R.A, Patel, D.J. | | Deposit date: | 1996-06-12 | | Release date: | 1996-12-23 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Molecular recognition in the bovine immunodeficiency virus Tat peptide-TAR RNA complex.
Chem.Biol., 2, 1995
|
|
135D
 
 | |
136D
 
 | |
8T8E
 
 | | cryoEM structure of Smc5/6 5mer | | Descriptor: | DNA repair protein KRE29, Non-structural maintenance of chromosome element 5, Structural maintenance of chromosomes protein 6 | | Authors: | Yu, Y, Patel, D.J. | | Deposit date: | 2023-06-22 | | Release date: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for Nse5-6 mediated regulation of Smc5/6 functions.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8T8F
 
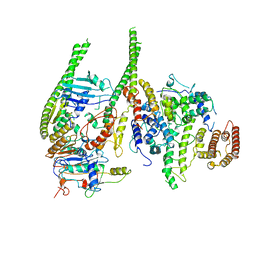 | | Smc5/6 8mer | | Descriptor: | DNA repair protein KRE29, Non-structural maintenance of chromosome element 4, Non-structural maintenance of chromosome element 5, ... | | Authors: | Yu, Y, Patel, D.J. | | Deposit date: | 2023-06-22 | | Release date: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Molecular basis for Nse5-6 mediated regulation of Smc5/6 functions.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8T64
 
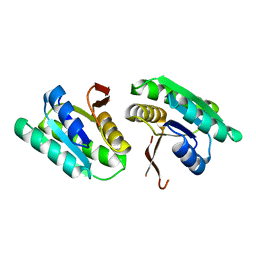 | | Apo Cam1(42-206) | | Descriptor: | Cam1 | | Authors: | Yu, Y, Patel, D.J. | | Deposit date: | 2023-06-15 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The CRISPR effector Cam1 mediates membrane depolarization for phage defence.
Nature, 625, 2024
|
|
8T65
 
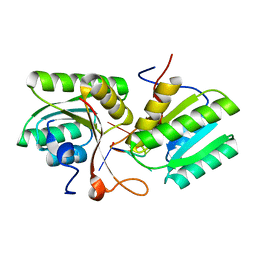 | | cA4 bound Cam1 | | Descriptor: | Cam1, RNA (5'-R(P*AP*AP*AP*A)-3') | | Authors: | Yu, Y, Patel, D.J. | | Deposit date: | 2023-06-15 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The CRISPR effector Cam1 mediates membrane depolarization for phage defence.
Nature, 625, 2024
|
|
8T66
 
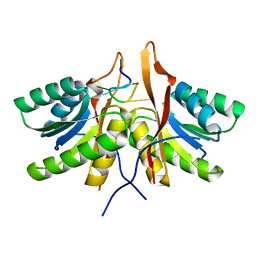 | | cA6 bound Cam1 | | Descriptor: | Cam1, RNA (5'-R(P*AP*AP*AP*AP*A)-3') | | Authors: | Yu, Y, Patel, D.J. | | Deposit date: | 2023-06-15 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The CRISPR effector Cam1 mediates membrane depolarization for phage defence.
Nature, 625, 2024
|
|
146D
 
 | | SOLUTION STRUCTURE OF THE MITHRAMYCIN DIMER-DNA COMPLEX | | Descriptor: | 1,2-HYDRO-1-OXY-3,4-HYDRO-3-(1-METHOXY-2-OXY-3,4-DIHYDROXYPENTYL)-8,9-DIHYROXY-7-METHYLANTHRACENE, 2,6-dideoxy-3-C-methyl-beta-D-ribo-hexopyranose-(1-3)-2,6-dideoxy-beta-D-galactopyranose-(1-3)-beta-D-Olivopyranose, DNA (5'-D(*TP*CP*GP*CP*GP*A)-3'), ... | | Authors: | Sastry, M, Patel, D.J. | | Deposit date: | 1993-11-09 | | Release date: | 1995-03-31 | | Last modified: | 2022-01-19 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the mithramycin dimer-DNA complex.
Biochemistry, 32, 1993
|
|
107D
 
 | | SOLUTION STRUCTURE OF THE COVALENT DUOCARMYCIN A-DNA DUPLEX COMPLEX | | Descriptor: | 4-HYDROXY-2,8-DIMETHYL-1-OXO-6-(4,5,6-TRIMETHOXY-1H-INDOLE-2-CARBONYL)-1,2,3,6,7,8-HEXAHYDRO-3,6-DIAZA-AS-INDACENE-2-CARBOXYLIC ACID METHYL ESTER, DNA (5'-D(*CP*CP*TP*TP*TP*TP*C)-3'), DNA (5'-D(*GP*AP*AP*AP*AP*GP*G)-3') | | Authors: | Lin, C.H, Patel, D.J. | | Deposit date: | 1995-01-17 | | Release date: | 1995-05-08 | | Last modified: | 2024-03-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the covalent duocarmycin A-DNA duplex complex.
J.Mol.Biol., 248, 1995
|
|
143D
 
 | |
